Abstract
Purpose
Bacteria of the genus Bartonella are obligate parasites of vertebrates. Their distribution range covers almost the entire world, from the Americas to Europe and Asia. Many Bartonella species use rodents as reservoirs, and while much is known about Bartonella infection of rodents in central Europe, its extent is poorly understood in Eastern Europe.
Methods
The present study examines five rodent species (Apodemus flavicollis, Myodes glareolus, Microtus arvalis, Apodemus agrarius, Apodemus sylvaticus) in the Chernobyl Exclusion Zone in Ukraine. Total of 36 small mammals were captured in September 2017.
Results
The overall prevalence of Bartonella spp. was 38.9% (14/36) in rodents. Obtained four sequences from Apodemus flavicollis, were identical to Bartonella grahamii and B. taylorii.
Conclusion
This is the first report to confirm the presence of Bartonella spp. in rodents in the Chernobyl Exclusion Zone, Ukraine by molecular methods. The sequences show similarity to Bartonella strains occurring in Europe.
Similar content being viewed by others
Introduction
The Chernobyl Exclusion Zone (CEZ) was established around the site of the 1986 disaster at the Chernobyl nuclear power plant. The area around the nuclear power plant was irradiated. Since this time, the inhabitants were resettled to another part of Ukraine and wild animals were allowed to return to the abandoned by people areas. The direct consequences of radioactive pollution, as well as the evacuation of the human population and cessation of agriculture and forest management resulted in significant ecological changes, such as the spontaneous restitution of the original natural habitats, the plant and animal populations, and their interdependences. The wild mammal population is now numerous and strong. Many aspects of the consequences of radionuclide contamination of wild animals have been studied during the short time since the catastrophe. For instance, Frantsevich [13] examines radionuclide accumulation in the soil and by different species of animals, Møller and Mousseau [29] report the effects of radiation on cytogenetics and mutation, and Møller et al. [30] examine the frequency of abnormalities potentially caused by such pollution. The increase of micromammalian parasite complexes and their biodiversity have also been examined [25, 34]. However, the parasite fauna of wild animals, including those of the small rodents in Chernobyl Exclusion Zone, is poorly understood, as are the observations of arthropod-borne pathogens [19]. The genus Bartonella comprises small, Gram-negative bacteria which act as obligate intracellular parasites of vertebrates. More than 30 species, as well as three subspecies, have been described [3, 7, 15]. Many of Bartonella species have been associated with emerging diseases, the symptoms of which have a broad spectrum of clinical syndromes ranging from cat-scratch diseases to potentially fatal conditions, such as endocarditis [1, 8, 9, 20,21,22, 43, 44]. Some Bartonella species, including Bartonella elizabethae, Bartonella tribocorum, Bartonella grahamii, Bartonella vinsonii subsp. arupensis and Bartonella washoensis, are causative agents of human infections [9, 21, 22, 43], and some researchers consider these bacteria to be host-specific, e.g. B. elizabethae and B. tribocorum are specific to Rattus spp. [3]. Bartonella species are transmitted by blood-sucking arthropods, such as sandflies, lice, fleas [6, 16, 31, 39]; in addition, Bartonella spp. have also been found in keds: Lipoptena cervi, Melophagus ovinus [11, 12, 40]. Bartonella species have also been common in selected rodents, ruminants and carnivores; the prevalence of infection in these animals group may be very high [6, 18] and strongly depends on the season [18]. Many animal-associated Bartonella species have been described and can be a threat to people. A high prevalence of Bartonella spp. has been observed in rodents in Asia 51%, in America 42% and in Africa 29% [2, 23, 28, 42]. Their prevalence in rodents in Central Europe ranges from 3.3 to 65.8% [18, 24, 26, 37, 41]. In European rodents, four species have been observed to be the most widespread: Bartonella grahamii, B. taylori, B. birtlesii and B. rochalimae. In Central Europe, the prevalence of Bartonella infection in small rodents is quite well known. Studies report a 65.8% prevalence in Germany, 9.0% in Slovenia, 3.3% in Croatia, 23.7% in Lithuania and 28% in Poland [18, 24, 26, 37, 41]. Although some reports exist of Bartonella infection in rodents in Eastern Europe, our understanding of their distribution remains incomplete. Therefore, the aim of this study was to analyse the occurrence of Bartonella species in rodents living near the abandoned city in the Chernobyl Exclusion Zone, Ukraine.
Materials and Methods
Study Area
The CEZ is located c. 200 km N of Kiev, Ukraine (51° N; 30.005° E), 123 m above sea level. This zone cover an area of 2600 km2 and falls entirely within the Polesie Lowland, Russian Plain. The climate in the CEZ is humid, with relatively mild winters and warm summers. The mean annual temperature is 5–7 °C, with a mean temperature in July of 18 °C (max. 32 °C), and in January of − 6 °C (min. − 25 °C). The annual precipitation ranges from 550 to 750 mm. The snow cover lasts on average c. 50 days per year. The mean depth of the snow cover is 12–13 cm [5].
The CEZ is surrounded by metal fencing 2 m high. Approximately c. 50 peasants also are still living in the CEZ; only few other persons have constant access to this area. Before the nuclear disaster, the CEZ consisted of farmland with patchy areas of forest. Currently, ca. 60% of the area is covered with forests, 50% of which is pine forest and remaining consist of abandoned arable grounds, meadows, pastures and human settlements.
Data Collection
Rodents were collected to the 3.2 km west of Chernobyl city (51°17′0′′ N; 30°13′2′′ E) in September 2017, using Sherman live-traps. The traps were checked two times per day. The captured mammals were transported in their traps to the laboratory for examination. The autopsies were carried out under terminal isoflurane anesthesia. Blood was collected for smear samples and spleen samples were taken. These were stored at − 20 °C and transported to the laboratory. The following animals were captured, in order of quantity: bank vole (Myodes glareolus)—13 animals, 4 m/9f (36.1% of total), yellow-necked mice (Apodemus flavicollis)—12 animals, 4 m/8f (33.3%), and striped field mice (Apodemus agrarius)—seven animals, 4 m/3f (19.44%). Rarer species included the common vole (Microtus arvalis)—three animals, 2 m/1f (8.3%), and one male wood mouse (Apodemus sylvaticus).
DNA Isolation and PCR Amplification
The DNA was isolated from spleen using the AX Tissue Mini Kit (A&A Biotechnology, Gdynia, Poland) according to the manufacturer’s protocol. For the PCR test, the primers rpoR and rpoF were used to detect Bartonella spp. These primers (1400F and 2300R) amplify an 850 bp fragment of the rpoB gene [35]. The PCR test was conducted according to Renesto et al. [35]. The PCR reactions were conducted in a 50 µl reaction mixture containing 2 µl of DNA template, 0.5 U (0.1 µl) of RUN Taq polymerase (A&A Biotechnology, Gdynia, Poland), 1 µl of dNTPs (10 mM), 0.5 µl of each primer (20 mM), and 5 µl of 10 × Taq DNA polymerase buffer (pH 8.6, 25 mM MgCl2). In the negative control, nuclease-free water was added to the PCR mix instead of the tested DNA. Both reactions were performed using the DNA Engine PTC-200 Thermal Cycler (BioRad, Hercules, USA) according to the following program: initial denaturation was performed at 94 °C for 5 min, followed by 35 cycles of denaturation at 95 °C for 10 s, annealing at 60 °C for 10 s and extension at 72 °C for 60 s. The final extension was performed at 72 °C for 7 min and then kept at 10 °C. The PCR products were visualized on a 1.0% agarose gel stained with ethidium bromide. Visualization was performed using ChemiDoc, MP Lab software (Imagine, BioRad, Hercules, USA). The resulting product was compared using the Nova 100 bp DNA Ladder Novazym (Poznań, Poland). The PCR amplicons were purified using a QIAEX II Gel Extraction Kit (Qiagen, Hilden, Germany), sequenced in both directions by Genomed (Poland), and contiguous sequences assembled using ContigExpress, Vector NTI Advance 11.0 (Invitrogen Life Technologies, New York, USA). The derived sequences were submitted to the GenBank database under the accession numbers MH669401-MH669404. To the phylogenetic studies, there was used Bayesian inference (BI) analysis with MrBayes version 3.2 [17]. Analysis of partial rpoB gene sequence data was based on an alignment of 762 bp (254 amino acids) using GTR + I + G model. The GTR models were chosen on the basis of jModelTest version 2.1.4 [10, 14] using Akaike information criterion.
Results
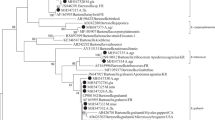
The molecular analysis revealed a higher prevalence of Bartonella spp. infection, with the total infection being 38.89% (14/36). In Apodemus flavicollis, the prevalence was 75% (9/12). The prevalence in Microtus arvalis (33.3%, 1/3) and Apodemus agrarius (28.57%, 2/7) was similar while the prevalence in Myodes glareolus was 15.38% (2/13). No infected individuals were found in the case of Apodemus sylvaticus. Two sequences (Bartonella sp. 1: MH669401-MH669402) obtained in this study share 100% similarity with B. taylorii isolated from Apodemus flavicollis from Lithuania (GenBank: MH547315) and B. taylorii strain M6 from France (GenBank: AF165995). In turn, two sequences (Bartonella sp. 2: MH669403-MH669404) showed 100% similarity with B. grahamii isolated from Myodes glareolus from Lithuania (MH547328) and 98.1% similarity with Bartonella sp. isolated from Clethrionomys rufocanus from Japan (AB290276) (Table 1; Fig. 1).
Phylogenetic tree of Bartonella spp., constructed by Bayesian inference (BI) analysis using MrBayes version 3.2. For BI codon analysis (nucmodel = codon), the GTR + I + G model was chosen based on jModelTest version 2.1.4 [10, 14] using Akaike Information Criterion. Analysis was run for 8,000,000 generations, with 2,000,000 generations discarded as ‘burn-in’. Hosts, country and GenBank accession numbers of origin are shown. Nodal support is indicated as Bayesian posterior probabilities. Sequence from Brucella melitensis (AY562179) was used as outgroup. Sequences generated in this study are show in bold
Discussion
Our findings indicate the presence of Bartonella species in rodents inhabited Chernobyl Exclusion Zone in Ukraine. The nearest studies of Bartonella infection in rodents have been conducted in Poland, Slovakia and Lithuania. In all of these countries, rodents were tested for bacteria genus Bartonella. The prevalence in all animals was 38.89% and it is comparable with results from Poland (22.4%) [33], and lower in Slovakia (64.8%) [38]. The prevalence among rodent species was varied. The prevalence in Apodemus flavicollis was high (75%), and was similar in another countries: Poland (66.6%) [18], Slovakia (63%) [38] and Lithuania (80%) [27]. The obtained prevalence of this bacterium in Myodes glareolus was 33.3%. This result was identical in Lithuania [27] and similar with result obtained in Poland (38.7% in the Mazurian Lakeland, 33.3% in Białowieża Forest) [4, 18] or from southwestern Slovakia (69.0%) [38]; however, they are higher than in western Slovakia (9.7%) [36]. In the case of Microtus arvalis, the prevalence of Bartonella spp. infection was 33.3%. This value is comparable with that obtained previously from Poland (27.7%) by Pawełczyk et al. [32], and in a PCR study in Mazuria, Poland (33.3%) by Paziewska et al. [33]. In contrast, this value was markedly higher than another result from Central Europe, where Šebek [36] identified a prevalence of 7.7% in western Slovakia. Compared to other rodent species, Apodemus agrarius has been poorly studied. Our studies show prevalence higher (28.57%) than in Slovakia [24] and Lithuania [27]. No infected individuals were found in the case of Apodemus sylvaticus, but we obtained only one animal. Two sequences (Bartonella sp. 1) obtained in this study are identical with B. taylorii isolated from Apodemus flavicollis from Lithuania (GenBank: MH547315) and B. taylorii strain M6 from France (GenBank: AF165995). This suggests that this Bartonella species occurs throughout Europe. Two sequences (Bartonella sp. 2) are identical with B. grahamii isolated from Myodes glareolus from Lithuania (MH547328) and 98.1% similarity with Bartonella sp. isolated from Clethrionomys rufocanus from Japan (AB290276). There is the first evidence of the infection of rodents in Chernobyl Exclusion Zone with Bartonella spp. The similarity of obtained rpoB gene sequences to isolates from Lithuania, as well from France, published in GenBank, indicates that rodents in CEZ are infected with common species of Bartonella. The follow-up studies are necessary to obtain the number of samples enough to statistical analysis, as well study conducted whole year, to describe the seasonal dynamic.
Our findings indicate that rodents in the Chernobyl Exclusion Zone are infected by strains of Bartonella common in Europe. The study presents the first record of bacteria genus Bartonella in rodents in the Chernobyl Exclusion Zone.
References
Anderson BE, Neuman MA (1997) Bartonella spp. as emerging human pathogens. Clin Microbiol Rev 10:203–219. https://doi.org/10.1128/cmr.10.2.203-219.1997
Bai Y, Calisher CH, Kosoy MY, Root JJ, Doty JB (2011) Persistent infection or successive reinfection of deer mice with Bartonella vinsonii subsp. arupiensis. Appl Environ Microbiol 77:1728–1731. https://doi.org/10.1128/aem.02203-10
Bai Y, Kosoy MY, Lerdthusnee K, Peruski LF, Richardson JH (2009) Prevalence and genetic heterogeneity of Bartonella strains cultured from rodents from 17 provinces in Thailand. Am J Trop Med Hyg 81:811–816. https://doi.org/10.4269/ajtmh.2009.09-0294
Bajer A, Welc-Falęciak R, Bednarska M, Alsarraf M, Behnke-Borowczyk J, Siński E, Behnke JM (2014) Long-term spatiotemporal stability and dynamic changes in the haemoparasite community of bank voles (Myodes glareolus) in NE Poland. Microb Ecol 68(2):196–211. https://doi.org/10.1007/s00248-014-0390-9
Barjachtar VG et al (2001) Chernobyl disaster 1996. Naukova Dumka Pre, Kyiv (In Russian)
Breitschwerdt EB, Kordick DL (2003) Bartonella infection in animals carriership reservoir potential, pathogenicity, and zoonotic potential for human infection. Clin Microbiol Rev 13:428–438. https://doi.org/10.1128/cmr.13.3.428-438.2000
Buffet JP, Kosoy M, Vayssier-Taussat M (2013) Natural history of Bartonella-infecting rodents in light of new knowledge on genomics, diversity and evolution. Future Microbiol 8:1117–1128. https://doi.org/10.2217/fmb.13.77
Chomel BB, Boulouis HJ, Breitschwerdt EB (2004) Cat scratch disease and other zoonotic Bartonella infections. J Am Vet Assoc 224:1270–1279. https://doi.org/10.2460/javma.2004.224.1270
Daly JS, Worthington MG, Brenner DJ, Moss CW, Hollis DG, Weyant RS, Steigerwalt AG, Weaver RE, Daneshvar MI, O’Connor SP (1993) Rochalimae elizabeathae sp. nov. isolated from a patient with endocarditis. J Clin Microbiol 31:872–881. https://doi.org/10.1128/jcm.31.4.872-881.1993
Darriba D, Taboada GL, Doallo R, Posada D (2012) jModelTest 2: more models, new heuristics and paralel computing. Nat Methods 9(8):772. https://doi.org/10.1038/nmeth.2109
Dehio C, Sauder U, Hiestand R (2004) Isolation of Bartonella schoenbchensis in Lipoptena cervi, a blood-sucking arthropod causing deer ked dermatitis. J Clin Microbiol 42:5320–5323. https://doi.org/10.1128/jcm.42.11.5320-5323.2004
Duan DY, Liu GH, Cheng TY, Wang YQ (2017) Microbial population analysis of the midgut of Melophagus ovinus via high-throughput sequencing. Parasit Vectors 9:382. https://doi.org/10.1186/s13071-017-2323-1
Frantsevich L (2006) Animal radioecology in the exclusion zone since the Chernobyl catastrophe. Zool Res 27:647–655
Guindon S, Gascuel OA (2003) Simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704. https://doi.org/10.1080/10635150390235520
Gutierrez R, Krasnov B, Morick D, Gottlieb Y, Khokhlova IS, Harrus S (2015) Bartonella infection in rodents and their flea ectoparasites: an overview. Vector Borne Zoonotic Dis 15:27–39. https://doi.org/10.1089/vbz.2014.1606
Halos L, Jamal T, Maillard R, Girard B, Guillot J, Chomel B, Vayssier-Taussat M, Boulouis HJ (2004) Role of Hippoboscidae flies as a potential vector of Bartonella spp. infecting wild domestic ruminants. Appl Environ Microbiol 70:6302–6305. https://doi.org/10.1128/aem.70.10.6302-6305.2004
Huelsenbeck JP, Ronquist F (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755. https://doi.org/10.1093/bioinformatics/17.8.754
Karbowiak G, Rychlik L, Nowakowski W, Wita I (2005) Natural infections of small mammals with blood parasites on the borderlands of boreal and temperate forest zone. Acta Theriol 50:31–42
Karbowiak G, Víchová B, Slivinska K, Werszko J, Didyk J, Peťko B, Stanko M, Akimov I (2014) The infection of questing Dermacentor reticulatus ticks with Babesia canis and Anaplasma phagocytophilum in the Chernobyl exclusion zone. Vet Parasitol 204:372–375. https://doi.org/10.1016/j.vetpar.2014.05.030
Kerhoff FT, Bergmans AM, van Der Zee A, Rothova A (1999) Demonstration of Bartonella grahamii DNA in ocular fluids of patient with neuroretinitis. J Clin Microbiol 37:4034–4038. https://doi.org/10.1128/jcm.37.12.4034-4038.1999
Kosoy M, Bai Y, Sheff K, Morway C, Baggett H, Maloney SA, Boonmar S, Bhengsri S, Dowell SF, Sitdhirasdr A, Lerdthusnee K, Richardson J, Peruski LF (2010) Identification of Bartonella infections in febrile human patient from Thailand and their potential animal reservoirs. Am J Trop Med Hyg 82:1140–1145. https://doi.org/10.4269/ajtmh.2010.09-0778
Kosoy M, Murray M, Gilmore RD Jr, Bai Y, Gage KL (2003) Bartonella strains from ground squirrels are identical to Bartonella washoensis isolated from a human patient. J Clin Microbiol 41:645–650. https://doi.org/10.1128/jcm.41.2.645-650.2003
Kosoy MY, Regnery RL, Tzianabos T, Marston EL, Jones DC, Green D, Maupin GO, Olson JG, Childs JE (1997) Distribution, diversity, and host specificity of Bartonella in rodents from the Southeastern United State. Am J Trop Med Hyg 57:578–588. https://doi.org/10.4269/ajtmh.1997.57.578
Kraljik J, Paziewska-Harris A, Miklisova D, Blaňarová L, Mošanský L, Bona M, Stanko M (2016) Genetic diversity of Bartonella genotypes found in the striped field mouse (Apodemus agarius) in central Europe. Parasitology 143:1437–1442. https://doi.org/10.1017/s0031182016000962
Labetskaya AG, Kiriyenko KM, Baydakova IV, Tishechkina IM (1997) An abundance and biodiversity of parasites from micromammalians in the evacuation zone of the Chernobyl Nuclear Station. Parasitologiya 31:391–396 (in Russian)
Lipatova I, Paulauskas A, Puraite I, Radzijevskaja J, Balciauskas L, Gedminas V (2015) Bartonella infection in small mammals and their ectoparasites in Lithuania. Microbes Infect 17:884–888. https://doi.org/10.1016/j.micinf.2015.08.013
Mardosaité-Busaitiené D, Radzijevskaja J, Balčiauskas L, Bratchikov M, Jurgelevičius V, Paulauskas A (2019) Prevalence and diversity of Bartonella species in small rodents from coastal and continental areas. Sci Rep 9(1):12349. https://doi.org/10.1038/s41598-019-48715-y
Meheretu Y, Leirs H, Welegerima K, Breno M, Tomas Z, Kidane D, Girmay K, de Belloeq JG (2013) Bartonella prevalence and genetic diversity in small mammals from Ethiopia. Vector Borne Zoonotic Dis 13:164–175. https://doi.org/10.1089/vbz.2012.1004
Møller AP, Mousseau TA (2006) Biological consequences of Chernobyl: 20 years on. Trends Ecol Evol 21:200–207. https://doi.org/10.1016/j.tree.2006.01.008
Møller AP, Mousseau TA, de Lope F, Saino N (2007) Elevated frequency of abnormalities in barn swallows from Chernobyl. Biol Lett 3:414–417. https://doi.org/10.1098/rsbl.2007.0136
Müller A, Reiter M, Schötta AM, Stockinger H, Stanek G (2016) Detection of Bartonella spp. in Ixodes ricinus ticks and Bartonella seroprevaence in human population. Ticks Tick Borne Dis 7:763–767. https://doi.org/10.1016/j.ttbdis.2016.03.009
Pawelczyk A, Bajer A, Behnke JM, Gilbert FS, Sinski E (2004) Factors affecting the component community structure of haemoparasites in common voles (Microtus arvalis) from the Mazury Lake District region of Poland. Parasitology 92:270–284. https://doi.org/10.1007/s00436-003-1040-1
Paziewska A, Harris PD, Zwolińska L, Bajer A, Siński E (2011) Recombination within and between species of the alpha proteobacterium Bartonella infecting rodents. Microb Ecol 61:134–145. https://doi.org/10.1007/s00248-010-9735-1
Pelgunov AN (2005) The parasites and parasitic systems in radiation biocenosis: the Chernobyl accident zone. Nauka, Moskow, p 207
Renesto P, Gouvernet J, Drancourt M, Roux V, Raoult D (2001) Use of rpoB analysis for detection and identification of Bartonella species. J Clin Microbiol 3:430–437. https://doi.org/10.1128/jcm.39.2.430-437.2001
Šebek Z (1975) Blutparasiten der wildlebenden Kleinsäuger in der Tschechoslowakei. Folia Parasitologica (Praha) 22:11–20
Silaghi C, Pfeffer M, Kiefer D, Kiefer M, Obiegala A (2016) Bartonella, rodents, fleas and ticks: a molecular field study on host-vector-pathogen association in Saxony, Germany. Microb Ecol 72:965–974. https://doi.org/10.1007/s00248-016-0787-8
Spitalska E, Minichova L, Kocianova E, Škultety L, Hamisikova Z, Slovak M, Kazimirova M (2017) Diversity and prevalence of Bartonella species in small mammals from Slovakia, Central Europe. Parasitol Res 116:3087–3095. https://doi.org/10.1007/s00436-017-5620-x
Sytykiewicz H, Karbowiak G, Werszko J, Czerniewicz P, Sprawka I, Mitrus J (2012) Molecular screening for Bartonella henselae and Borrelia burgdorferi sensu lato co-existence within Ixodes ricinus populations in central ane eastern parts of Poland. Ann Agric Environ Med 19:451–456
Szewczyk T, Werszko J, Steiner-Bogdaszewska Ż, Jeżewski W, Laskowski Z, Karbowiak G (2017) Molecular detection of Bartonella spp. in deer ked (Lipoptena cervi) in Poland. Parasit Vectors 10:487. https://doi.org/10.1186/s13071-012-2413-0
Tadin A, Tokarz R, Markotić A, Margaletić J, Turk N, Habuš J, Svoboda P, Vucelja M, Desai A, Jain K, Lipkin WI (2016) Molecular survey of zoonotic agents in rodents and other small mammals in Croatia. Am J Trop Med Hyg 94:466–473. https://doi.org/10.4269/ajtmh.15-0517
Tsai YL, Chuang ST, Chang CC, Kass PH, Chomel BB (2010) Bartonella species in small mammals and their ectoparasites in Taiwan. Am J Trop Med Hyg 83:917–923. https://doi.org/10.4269/ajtmh.2010.10-0083
Welch DF, Carroll KC, Hofmeister EK, Persing DH, Robinson DA, Steigerwalt AG, Brenner DJ (1999) Isolation of a new subspecies, Bartonella vinsonii subsp. arupensis, from a cattle rancher: identity with isolates found in conjunction with Borrelia burgdorferi and Babesia microti among naturally infected mice. J Clin Microbiol 37:2598–2601. https://doi.org/10.1128/jcm.37.8.2598-2601.1999
Welch DF, Pickett DA, Slater LN, Steigerwalt AG, Brenner DJ (1992) Rochalimaea henselae sp. nov., a cause of septicemia, bacillary angiomatosis, and parenchymal bacillary peliosis. J Clin Microbiol 30:275–280. https://doi.org/10.1128/jcm.30.2.275-280.1992
Acknowledgements
We would like to thank Mr. Alexander Borovskyi for help in collecting the field data, Dr. Sergey Gashchak and Dr. Igor Chizevsky from the Chernobyl Center for Nuclear Safety, Radioactive Waste and Radioecology for giving traps for our study.
Author information
Authors and Affiliations
Contributions
TS planned and organized the study. JW and KS collected samples. JW extracted DNA. TS and ZL performed PCR, sequencing and analyzed sequence data. TS and JW drafted the manuscript, and wrote the final version together with GK and KS. All authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflicts of interest.
Research Involving Human Participants and/or Animals
The permission of I.I. Schmalhausen Institute of Zoology of National Academy of Sciences of Ukraine; the procedures conducted according with the Directive 2010/63/EU of the European Union.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Szewczyk, T., Werszko, J., Slivinska, K. et al. Molecular Detection of Bartonella spp. in Rodents in Chernobyl Exclusion Zone, Ukraine. Acta Parasit. 66, 222–227 (2021). https://doi.org/10.1007/s11686-020-00276-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11686-020-00276-1