Abstract
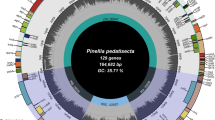
Populus alba is a foundation species in evolutionary and ecological studies in the northern hemisphere. In this study, the chloroplast genome and gene map of P. alba were constructed. The P. alba chloroplast genome is 156,505 bp in length comprising a large single-copy region, two inverted repeat regions and a small single-copy region. The genome contains 131 genes, including 86 protein-coding genes (77 PCG species), eight ribosomal RNA genes (four rRNA species) and 37 transfer RNA genes (30 tRNA species). Phylogenetic analysis indicates that all Populus chloroplast genome sequences are clustered together and divided into three large branches. Among reported Populus chloroplast genomes, the leuce section formed monophyletic, indicating that all Populus spp. have a common maternal ancestor. P. rotundifolia and P. tremula are closely related and are sisters to P. davidiana. P. alba is closely related to P. adenopoda. Population genetic research in ecology and evolution may be easily developed through chloroplast genomes as they are conserved. This research will benefit future studies related to Populus, one of the world’s most ecologically and economically important genera.



Similar content being viewed by others
References
Bergsten J (2005) A review of long-branch attraction. Cladistics 21(2):163–193
Corriveau JL, Coleman AW (1988) Rapid screening method to detect potential biparental inheritance of plastid DNA and results for over 200 angiosperm species. Am J Bot 75(10):1443–1458
Doyle J (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19(1):11–15
Du SH, Wang ZS, Ingvarsson P, Wang DS, Wang JH, Wu ZQ, Tembrock LR, Zhang JG (2015) Multilocus analysis of nucleotide variation and speciation in three closely related Populus (Salicaceae) species. Mol Ecol 24(19):4994–5005
He Y, Xiao HT, Deng C, Xiong L, Yang J, Peng C (2016) The complete chloroplast genome sequences of the medicinal plant Pogostemon cablin. Int J Mol Sci 17(6):820
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30(4):772–780
Koo YB, Yeo JK, Woo KS, Kim TS (2007) Selection of superior clones by stability analysis of growth performance in Populus davidiana dode at age 12. Silvae Genet 56(3):93–101
Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9(4):357–359
Lee KM, Yong YK, Hyun JO (2011) Genetic variation in populations of Populus davidiana Dode based on microsatellite marker analysis. Genes Genom 33(2):163–171
Lohse M, Bolger AM, Nagel A, Fernie AR, Lunn JE, Stitt M, Usadel B (2012) RobiNA: a user-friendly, integrated software solution for RNA-Seq-based transcriptomics. Nucleic Acids Res 40(Web Server issue):622–627
Ohyama K, Fukuzawa H, Kohchi T, Shirai H, Sano T, Sano S, Umesono K, Shiki Y, Takeuchi M, Chang Z, Aota S-I, Inokuchi H, Ozeki H (1986) Chloroplast gene organization deduced from complete sequence of liverwort Marchantia polymorpha chloroplast DNA. Nature 322(6079):572–574
Shinozaki K, Ohme M, Tanaka M, Wakasugi T, Hayashida N, Matsubayashi T, Zaita N, Chunwongse J, Obokata J, Yamaguchi-Shinozaki K, Ohto C, Torazawa K, Meng B-Y, Sugita M, Deno H, Kamogashira T, Yamada K, Kusuda J, Takaiwa F, Kato A, Tohdoh N, Shimada H, Sugiura M (1986) The complete nucleotide sequence of the tobacco chloroplast genome: its gene organization and expression. EMBO J 4(3):2043–2049
Wang ZS, Du SH, Dayanandan S, Wang DS, Zeng YF, Zhang JG (2014) Phylogeny reconstruction and hybrid analysis of Populus (Salicaceae) based on nucleotide sequences of multiple single-copy nuclear genes and plastid fragments. PLoS ONE 9(8):e103645
Wicke S, Schneeweiss GM, dePamphilis CW, Müller KF, Quandt D (2011) The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol Biol 76(3–5):273–297
Wu ZQ, Ge S (2012) The phylogeny of the BEP clade in grasses revisited: evidence from the whole-genome sequences of chloroplasts. Mol Phylogenet Evol 62(1):573–578
Acknowledgements
We thank Tian Yang for her suggestions on data analysis.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Project funding: The work was supported by the Fundamental Research Funds of the Chinese Academy of Forestry (No. CAFYBB2017ZX001-1) and the National Natural Science Foundation of China (No. 31470665).
The online version is available at http://www.springerlink.com
Corresponding editor: Tao Xu.
Rights and permissions
About this article
Cite this article
Hou, Z., Wang, Z. & Zhang, J. The complete chloroplast genomic landscape and phylogenetic analyses of Populus alba L.. J. For. Res. 31, 1875–1879 (2020). https://doi.org/10.1007/s11676-019-00953-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11676-019-00953-6