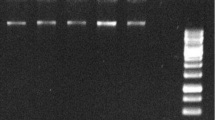
Abstract
An orthogonal design was used to optimize SSR-PCR amplification system using Panax ginseng genomic DNA as template. Four levels of five factors (DNA template, Taq DNA polymerase, Mg2+, primer, and dNTP) and annealing temperature have been tested separately in this system. The results demonstrated the reaction efficiency was affected by these factors. Based on the results, a stable, productive and reproducible PCR system and cycling program for amplifying a ginseng SSR locus were obtained: 20 µL system containing 1.0 U Taq DNA polymerase, 2.0 mmol·L−1 Mg2+, 0.2 mmol·L−1 dNTPs, 0.3 µmol·L−1 SSR primer, 60 ng·µL−1 DNA template, performed with a program of 94°C for 5 min, 94°C for 30 s, annealing at 56.3°C for 30 s, 72°C for 1 min, 37 cycles, finishing at 72°C for 7 min, and storing at 4°C.
Similar content being viewed by others
References
Dieffenbach, C.W., Dveksler, G.S. 2002. PCR Primer: A laboratory manual. Beijing: Science Press, p140–141.
He Zhengwen, Liu Yunsheng, Chen Lihua, et al. 1998. Orthogonal design-direct analysis for PCR optimization. Bulletin of Human Medical University, 23(4): 403–404. (in Chinese)
Jiang Yuxin, Shi Sixin, Zhang Zhie, et al. 1998. RAPD Analysis of genomic DNA in cultivars of Panax ginseng C.A.Mey. and Panax quinquefolius Linn., Special Wild Economic Animal and Plant Research, 1: 5–9. (in Chinese)
Katti, M.V., Ranjekar, P.K., Gupta, V.S. 2001. Differential distribution of simple sequence repeats in eukaryotic genome sequences. Mol. Biol. Evol., 18: 1161–1167.
Li Jinbo, Fang Xuanjun, Yang Guocai, et al. 2005. SSR fingerprinting the parents of two-line hybrid rice and its application in purity identification of hybrid seeds. Hybrid Rice, 20(2): 50–53. (in Chinese)
Litt, M., Luty, J.A. 1989. A hypervariable microsatellite revealed by in vitro amplification of a dinucleotide repeat within the cardiac muscle actin gene. Am. J. Hum. Genet, 44(3): 397–401.
Ma Xiaojun, Wang Xiaoquan, Xiao Peigen, et al. 2000. A study on AFLP fingerprinting of land races of Panax ginseng. China Journal of Chinese Materia Medica, 25(12): 707–710. (in Chinese)
Qiao Yushan, Zhang Zhen, Shen Zhijun, et al. 2004. Establishment of simple sequence repeat reaction system in Prunus salicina. Plant Physiology Communications, 40(1): 83–86. (in Chinese)
Scott O R, Arnold J B. 1985. Extraction of DNA from milligram amounts of fresh, herbarium and mummified plant tissues. Plant Molecular Biology, 5: 69–76.
Tautz, D. 1989. Hypervariablity of simple sequences as a general source for polymorphic DNA markers. Nucleic Acids Res., 17: 6463–6471.
Tóth, G., Gáspári, Z., Jurka, J. 2000. Microastellites in different eukaryotic genomes: survey and analysis. Genome Research, 10: 967–981.
Wang Biao, Qiu Lijuan, 2002. Current advance of simple sequence repeats in soybean. Chinese Bulletin of Botany, 19(1): 44–48. (in Chinese)
Wang Qiong, Cheng Zhou, Zhang Lu, et al. 2004. Distinguishing wild Panax ginseng from cultivated Panax ginseng based on direct amplification of length polymorphism (DALP) analysis. Journal of Fudan University (Natural Science), 43(6): 1030–1034. (in Chinese)
Xia Xueyan, Chang Jinhua, Luo Yaowu, et al. 2004. The optimization of SSR technique system on greenbug resistance in sorghum. Journal of Agricultural University of Hebei, 27(3): 21–24. (in Chinese)
Yuan Zhifa, Zhou Jingyu. 2000. Experimental design and analysis. Beijing: Higher Education Press, p467. (in Chinese)
Zhang Na, Yang Wenxiang, Meng Qingfang, et al. 2005. Study on optimization of wheat SSR reaction system and condition. Journal of Agricultural University of Hebei, 28(6): 5–8. (in Chinese)
Zhang Zhiqing, Zheng Youliang, Wei Yuming, et al. 2002. Analysis on genetic diversity among Sichuan wheat cultivars based on SSR markers. Acta Tritical Crops, 22(2): 5–9. (in Chinese)
Zhong Changsong, Xu Liyuan, Yu Guirong, et al. 2006. Genetic relationship among 20 special maize inbreds studied by SSR markers. Journal of Maize Sciences, 14(4): 43–46. (in Chinese)
Zhou Yanqing. 2005. Application of DNA molecule tag in plant research. Beijing: Chemical Industry Press, pp.133.
Author information
Authors and Affiliations
Additional information
Foundation project: This research was supported by Department of Wildlife Conservation, State Forestry Administration, P. R. China.
Electronic supplementary material is available in the online version of this article at http://dxdoi.org/10.1007/s11676-007-0006-z
Biography: YANG Tian-tian (1979–), female, postgraduate student of School of Forestry, Northeast Forestry University, Harbin 150040, P. R. China.
Rights and permissions
About this article
Cite this article
Tian-tian, Y., Li-qiang, M. & Jun, W. Optimizing SSR-PCR system of Panax ginseng by orthogonal design. J. of For. Res. 18, 31–34 (2007). https://doi.org/10.1007/s11676-007-0006-z
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/s11676-007-0006-z