Abstract
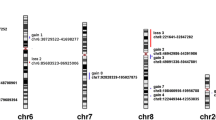
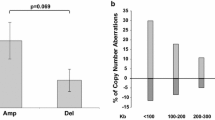
Objective: To detect genetic alterations in nasopharyngeal carcinoma (NPC) in Cantonese, the population with the highest incidence of NPC, and to correlate the findings with clinical staging. Methods: Comparative genomic hybridization (CGH) was performed on 35 primary nasopharyngeal carcinomas and a nonparametric x2 test was used to analyze relationship between chromosome changes and clinical staging. Results: The identified common chromosomal alterations in NPC included gain of chromosomes 12q (21 cases, 60%), 4q (19cases, 43%), 3q (18 cases, 51%), 1q (15 cases, 43%),8q (14 cases, 40%), and 2q (12 cases, 30%). The most frequently detected loss of chromosomal materials involved chromosome 1p (24 cases, 69%), chromosome 3p (21 cases, 60%), 11q (20 cases, 57%), 14q (18 cases, 51%), 16q (14 cases, 40%), 13(12 cases, 34%), and 9p(11 cases, 31%). The high frequency (>50%) 4q gain and 1p loss were novel findings. Compared by nonparametric x2 test, gains on 12q and 8q were found mainly in stages III/IV and there were significant differences between two clinical stage groups (stages I/II vs stages III/IV). Conclusions: Current analysis has revealed a comprehensive profile of the chromosomal regions showing DNA copy number changes, which may harbor oncogenes or tumor suppressor genes involved in the development of primary NPC.
Similar content being viewed by others
References
Hildesheim A and Levine PH. Etiology of nasopharyngeal carcinoma: a review[J]. Epidemiol Rev 1993; 15:466.
2. Kallioniemi A, Kallioniemi OP, Sudar D, et al. Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors[J]. Science 1992; 258:818.
Chu LW, Pettaway CA, Liang JC. Genetic abnormalities specifically associated with varying metastatic potential of prostate cancer cell lines as detected by comparative genomic hybridization[J]. Cancer Genet Cytogenet 2001; 127: 161.
Helou K, Walentinsson A, Beckmann B, et al. Analysis of genetic changes in rat endometrial carcinomas by means of comparative genomic hybridization[J]. Cancer Genet Cytogenet 2001; 127: 118.
Soulier J, Pierron G, Vecchione D, et al. A complex pattern of recurrent chromosomal losses and gains in T-cell prolymphocytic leukemia[J]. Genes Chromosomes Cancer 2001; 31: 248.
Tsukasaki K, Krebs J, Nagai K, et al. Comparative genomic hybridization analysis in adult T-cell leukemia/lymphoma: correlation with clinical course[J]. Blood 2001; 97: 3875.
Sowery RD, Jensen C, Morrison KB, et al. Comparative genomic hybridization detects multiple chromosomal amplifications and deletions in undifferentiated embryonic sarcoma of the liver[J]. Cancer Genet Cytogenet. 2001; 126: 128.
Kim MH, Stewart J, Devlin C, et al. The application of comparative genomic hybridization as an additional tool in the chromosome analysis of acute myeloid leukemia and myelodysplastic syndromes[J]. Cancer Genet Cytogenet 2001; 126: 26.
Chen YJ, Ko JY, Chen PJ, et al. Chromosomal aberrations in nasopharyngeal carcinoma analyzed by comparative genomic hybridization[J]. Genes Chromosomes Cancer 1999; 25:169.
Fan CS, Wong N, Leung SF, et al. Frequent c-myc and Int-2 overrepresentations in nasopharyngeal carcinoma[J]. Hum Pathol. 2000; 31: 169.
Fang Y, Guan X, Guo Y, et al. Analysis of genetic alterations in primary nasopharyngeal carcinoma by comparative genomic hybridization[J]. Genes Chromosomes Cancer 2001; 30:254.
Hui AB, Lo KW, Leung SF, et al. Detection of recurrent chromosomal gains and losses in primary nasopharyngeal carcinoma by comparative genomic hybridization[J]. Intl J Cancer 1999; 82: 498.
Qian W, Hu LF, Wang Y, et al. Infrequent MDM2 gene amplification and absence of gross WALF gene alterations in nasopharyngeal carcinoma[J]. Europ J Cancer Oral Oncol 1995; 31B: 328.
Simons JW. Genetic, epigenetic, dysgenetic and non-genetic mechanisms in tumorigenesis. II. Further delineation of the rate limiting step[J]. Anticancer Res 1999; 19(6A):4781.
Yan J, Fang Y, Jiao Y, et al. Detection of c-myc amplification in primary nasopharyngeal carcinoma by interphase FISH[J]. Chin J Med Genet 2001; 18: 222.
Author information
Authors and Affiliations
Corresponding author
Additional information
Foundation item: This work was supported by the “973” National Key Basic Research Program of China (No.G1998051202) and the National Scientific Research Foundation for Excellent Young Scientist of China(type B) (No. 39825511).
Biography: Yan Jian,(1962–), doctor of medicine, attending physician, works at Cancer Center, Sun Yat-sen University of Medical Sciences, majors in oncology.
Rights and permissions
About this article
Cite this article
Yan, J., Fang, Y., Liang, Q. et al. DNA copy profile in nasopharyngeal carcinoma and its correlation with clinical staging. Chin. J. Cancer Res. 13, 276–279 (2001). https://doi.org/10.1007/s11670-001-0047-3
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/s11670-001-0047-3