Abstract
Purpose
To demonstrate how artificial intelligence (AI) can expand radiologists’ capacity, we visualized the features of invasive ductal carcinomas (IDCs) that our algorithm, developed and validated for basic pathological classification on mammograms, had focused on.
Materials and methods
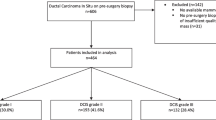
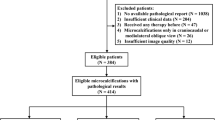
IDC datasets were built using mammograms from patients diagnosed with IDCs from January 2006 to December 2017. The developing dataset was used to train and validate a VGG-16 deep learning (DL) network. The true positives (TPs) and accuracy of the algorithm were externally evaluated using the test dataset. A visualization technique was applied to the algorithm to determine which malignant findings on mammograms were revealed.
Results
The datasets were split into a developing dataset (988 images) and a test dataset (131 images). The proposed algorithm diagnosed 62 TPs with an accuracy of 0.61–0.70. The visualization of features on the mammograms revealed that the tubule forming, solid, and scirrhous types of IDCs exhibited visible features on the surroundings, corners of the masses, and architectural distortions, respectively.
Conclusion
We successfully showed that features isolated by a DL-based algorithm trained to classify IDCs were indeed those known to be associated with each pathology. Thus, using AI can expand the capacity of radiologists through the discovery of previously unknown findings.



Similar content being viewed by others
References
Silver D, Huang A, Maddison CJ, Guez A, Sifre L, Van Den Driessche G, et al. Mastering the game of Go with deep neural networks and tree search. Nature. 2016;529(7587):484–9. https://doi.org/10.1038/nature16961.
LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521(7553):436–44. https://doi.org/10.1038/nature14539.
Yasaka K, Akai H, Kunimatsu A, Kiryu S, Abe O. Deep learning with convolutional neural network in radiology. Jpn J Radiol. 2018;36(4):257–72. https://doi.org/10.1007/s11604-018-0726-3.
Tsuda H, Tsugawa K, Akiyama F, et al. Histological classification of breast tumors in the General Rules for Clinical and Pathological Recording of Breast Cancer (18th edition). Breast Cancer. 2020;27:309–21. https://doi.org/10.1007/s12282-020-01074-3.
Selvaraju RR, Cogswell M, Das A, Vedantam R, Parikh D, Batra D, editors. Grad-cam: Visual explanations from deep networks via gradient-based localization. In: Proceedings of the IEEE international conference on computer vision; 2017. pp. 618–626. https://doi.org/10.1109/ICCV.2017.74.
Collins GS, Reitsma JB, Altman DG, Moons KGM. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): the TRIPOD statement. BMJ. 2015;350:g7594. https://doi.org/10.1136/bmj.g7594.
Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556. 2014.
Zou L, Yu S, Meng T, Zhang Z, Liang X, Xie Y. A technical review of convolutional neural network-based mammographic breast cancer diagnosis. Comput Math Methods Med. 2019;2019:6509357.
Ma W, Zhao Y, Ji Y, Guo X, Jian X, Liu P, et al. Breast cancer molecular subtype prediction by mammographic radiomic features. Acad Radiol. 2019;26(2):196–201.
Goodfellow IJ, Shlens J, Szegedy C. Explaining and harnessing adversarial examples. arXiv preprint arXiv:1412.6572. 2014.
Ueda D, Shimazaki A, Miki Y. Technical and clinical overview of deep learning in radiology. Jpn J Radiol. 2019;37(1):15–33. https://doi.org/10.1007/s11604-018-0795-3.
Funding
This study was supported by Kaken JSPS KAKENHI Grant number JP18K15597.
Author information
Authors and Affiliations
Contributions
DU: prepared the development data and performed the data analysis; AY, ST, and YM: supervised the project; NO, TT, SN, SK, and TM: provided mammograms; AS, HT and TG: revised the manuscript.
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
About this article
Cite this article
Ueda, D., Yamamoto, A., Takashima, T. et al. Visualizing “featureless” regions on mammograms classified as invasive ductal carcinomas by a deep learning algorithm: the promise of AI support in radiology. Jpn J Radiol 39, 333–340 (2021). https://doi.org/10.1007/s11604-020-01070-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11604-020-01070-9