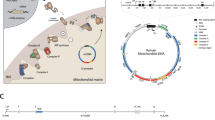
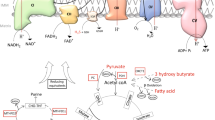
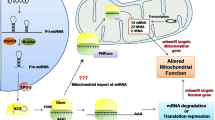
Abstract
The energy shift toward glycolysis is one of the hallmarks of cancer. Complex I is a vital enzyme complex necessary for oxidative phosphorylation. The mitochondrially encoded NADH: ubiquinone oxidoreductase core subunit 1 (MT-ND1) is the largest subunit coded by mitochondria of complex I. The present study summarizes the structure and biological function of MT-ND1. From databases and literature, the expressions and mutations of MT-ND1 in a variety of cancers have been reviewed. MT-ND1 may be a biomarker for cancer diagnosis and prognosis. It is also a potential target for cancer therapy.
Similar content being viewed by others
References
Naithani S, Saracco SA, Butler CA, et al. Interactions among COX1, COX2, and COX3 mRNA-specific translational activator proteins on the inner surface of the mitochondrial inner membrane of Saccharomyces cerevisiae. Mol Biol Cell, 2003,14(1):324–333
Lenka N, Vijayasarathy C, Mullick J, et al. Structural organization and transcription regulation of nuclear genes encoding the mammalian cytochrome c oxidase complex. Prog Nucleic Acid Res Mol Biol, 1998,61:309344
Gazizova, N, Rakhmatullina D, Minibayeva F. Effect of respiratory inhibitors on mitochondrial complexes and ADP/ATP translocators in the Triticum aestivum roots. Plant Physiol Biochem, 2020,151:601–607
Giulivi, C, Zhang YF, Omanska-Klusek A, et al. Mitochondrial dysfunction in autism. JAMA, 2010,304(21):2389–2396
Holzknecht M, Guerrero-Navarro L, Petit M, et al. The mitochondrial enzyme FAHD1 regulates complex II activity in breast cancer cells and is indispensable for basal BT-20 cells in vitro. FEBS Lett, 2022,596(21):2781–2794
Baracca A, Solaini G, Sgarbi G, et al. Severe impairment of complex I-driven adenosine triphosphate synthesis in leber hereditary optic neuropathy cybrids. Arch Neurol, 2005,62(5):730–736
Huoponen, K. Leber hereditary optic neuropathy: clinical and molecular genetic findings. Neurogenetics, 2001,3(3):119–125
Lin J, Zhao CB, Lu JH, et al. Novel mutations m.3959G>A and m.3995A>G in mitochondrial gene MT-ND1 associated with MELAS. Mitochondrial DNA, 2014,25(1):56–62
Lenaz G, Baracca A, Carelli V, et al. Bioenergetics of mitochondrial diseases associated with mtDNA mutations. Biochim Biophys Acta, 2004,1658(1–2):89–94
Mitchell AL, Elson JL, Howell N, et al. Sequence variation in mitochondrial complex I genes: mutation or polymorphism? J Med Genet, 2006,43(2):175–179
Simon DK, Friedman J, Breakefield XO, et al. A heteroplasmic mitochondrial complex I gene mutation in adult-onset dystonia. Neurogenetics, 2003,4(4):199–205
Jin X, Zhang J, Yi Q, et al. Leber’s Hereditary Optic Neuropathy Arising From the Synergy Between ND1 3635G>A Mutation and Mitochondrial YARS2 Mutations. Invest Ophthalmol Vis Sci, 2021,62(7):22
Lin Y, Xu X, Zhao D, et al. A novel m.11406 T > A mutation in mitochondrial ND4 gene causes MELAS syndrome. Mitochondrion, 2020,54:57–64
Cieslik M, Czapski GA, Wójtowicz S, et al. Alterations of Transcription of Genes Coding Anti-oxidative and Mitochondria-Related Proteins in Amyloid beta Toxicity: Relevance to Alzheimer’s Disease. Mol Neurobiol, 2020,57(3):1374–1388
Anderson WM, Fisher RR. The subunit structure of bovine heart mitochondrial transhydrogenase. Biochim Biophys Acta, 1981,635(1):194–199
Wallace DC. Mitochondrial DNA sequence variation in human evolution and disease. Proc Natl Acad Sci U S A, 1994,91(19):8739–8746
Montoya J, Lopez-Perez MJ. Mitochondrial heterogeneity in Aspergillus nidulans: in vivo protein biosynthetic activities of the mitochondrial populations. Rev Esp Fisiol, 1980,36(4):427–431
Ojala D, Montoya J, Attardi G. tRNA punctuation model of RNA processing in human mitochondria. Nature, 1981,290(5806):470–474
Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin, 2021,71(3):209–249
Huang X, Zhu X, Yu Y, et al. Dissecting miRNA signature in colorectal cancer progression and metastasis. Cancer Lett, 2021,501:66–82
Akouchekian M, Houshmand M, Akbari MH, et al. Analysis of mitochondrial ND1 gene in human colorectal cancer. J Res Med Sci, 2011,16(1):50–55
Kassem AM, El-Guendy N, Tantawy M, et al. Mutational hotspots in the mitochondrial D-loop region of cancerous and precancerous colorectal lesions in Egyptian patients. DNA Cell Biol, 2011,30(11):899–906
Yusnita Y, Norsiah MD, Rahman AJ. Mutations in mitochondrial NADH dehydrogenase subunit 1 (mtND1) gene in colorectal carcinoma. Malays J Pathol, 2010,32(2):103–110
Koshikawa N, Akimoto M, Hayashi JI, et al. Association of predicted pathogenic mutations in mitochondrial ND genes with distant metastasis in NSCLC and colon cancer. Sci Rep, 2017,7(1):15535
Weerts MJA, Timmermans EC, van de Stolpe A, et al. Tumor-Specific Mitochondrial DNA Variants Are Rarely Detected in Cell-Free DNA. Neoplasia, 2018,20(7):687–696
Xu Y, Zhou J, Yuan Q, et al. Quantitative detection of circulating MT-ND1 as a potential biomarker for colorectal cancer. Bosn J Basic Med Sci, 2021,21(5):577–586
Pinheiro M, Veiga I, Pinto C, et al. Mitochondrial genome alterations in rectal and sigmoid carcinomas. Cancer Lett, 2009,280(1):38–43
Habano W, Nakamura S, Sugai T. Microsatellite instability in the mitochondrial DNA of colorectal carcinomas: evidence for mismatch repair systems in mitochondrial genome. Oncogene, 1998,17(15):1931–1937
Wallace L, Mehrabi S, Bacanamwo M, et al. Expression of mitochondrial genes MT-ND1, MT-ND6, MT-CYB, MT-COI, MT-ATP6, and 12S/MT-RNR1 in colorectal adenopolyps. Tumour Biol, 2016,37(9):12465–12475
Perou CM, Sorlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature, 2000,406(6797):747–52
Jahani MM, Azimi Meibody A, Karimi T, et al. An A10398G mitochondrial DNA alteration is related to increased risk of breast cancer, and associates with Her2 positive receptor. Mitochondrial DNA A DNA Mapp Seq Anal, 2020,31(1):11–16
Thapa S, Lalrohlui F, Ghatak S, et al. Mitochondrial complex I and V gene polymorphisms associated with breast cancer in mizo-mongloid population. Breast Cancer, 2016,23(4):607–616
Parrella P, Xiao Y, Fliss M, et al. Detection of mitochondrial DNA mutations in primary breast cancer and fine-needle aspirates. Cancer Res, 2001,61(20):7623–7626
Grzybowska-Szatkowska L, Slaska B. Mitochondrial NADH dehydrogenase polymorphisms are associated with breast cancer in Poland. J Appl Genet, 2014,55(2):173–181
Srigley JR, Delahunt B, Eble JN, et al. The International Society of Urological Pathology (ISUP) Vancouver Classification of Renal Neoplasia. Am J Surg Pathol, 2013,37(10):1469–1489
Kim H, Komiyama T, Inomoto C, et al. Mutations in the Mitochondrial ND1 Gene Are Associated with Postoperative Prognosis of Localized Renal Cell Carcinoma. Int J Mol Sci, 2016,17(12):2049
Kim H, Komiyama T, Nitta M, et al. D-loop Mutations in Renal Cell Carcinoma Improve Predictive Accuracy for Cancer-Related Death by Integrating with Mutations in the NADH Dehydrogenase Subunit 1 Gene. Genes (Basel), 2019,10(12):998
Mayr JA, Meierhofer D, Zimmermann F, et al. Loss of complex I due to mitochondrial DNA mutations in renal oncocytoma. Clin Cancer Res, 2008,14(8):2270–2275
Schutte K, Bornschein J, Malfertheiner P. Hepatocellular carcinoma—epidemiological trends and risk factors. Dig Dis, 2009,27(2):80–92
Li W, Qi Y, Cui X, et al. Heteroplasmy and Copy Number Variations of Mitochondria in 88 Hepatocellular Carcinoma Individuals. J Cancer, 2017,8(19):4011–4017
Yin PH, Wu CC, Lin JC, et al. Somatic mutations of mitochondrial genome in hepatocellular carcinoma. Mitochondrion, 2010,10(2):174–182
Liu L, Tan D, Wong LJ. Somatic mutation detection in complete mitochondrial DNA of lung cancer patients. Zhongguo Fei Ai Za Zhi (Chinese), 2004,7(2):125–129
Wang L, Wang J, Jia E, et al. Plasma RNA sequencing of extracellular RNAs reveals potential biomarkers for non-small cell lung cancer. Clin Biochem, 2020,83:65–73
Maximo V, Sobrinho-Simoes M. Hurthle cell tumours of the thyroid. A review with emphasis on mitochondrial abnormalities with clinical relevance. Virchows Arch, 2000,437(2):107–115
Maxim V, Soares P, Lima J, et al. Mitochondrial DNA somatic mutations (point mutations and large deletions) and mitochondrial DNA variants in human thyroid pathology: a study with emphasis on Hurthle cell tumors. Am J Pathol, 2002,160(5):1857–1865
Xu B, Reznik E, Tuttle RM, et al. Outcome and molecular characteristics of non-invasive encapsulated follicular variant of papillary thyroid carcinoma with oncocytic features. Endocrine, 2019,64(1):97–108
Porcelli AM, Ghelli A, Ceccarelli C, et al. The genetic and metabolic signature of oncocytic transformation implicates HIF1alpha destabilization. Hum Mol Genet, 2010,19(6):1019–1032
Bonora E, Porcelli AM, Gasparre G, et al. Defective oxidative phosphorylation in thyroid oncocytic carcinoma is associated with pathogenic mitochondrial DNA mutations affecting complexes I and III. Cancer Res, 2006,66(12):6087–6096
Jiang Z, Bahr T, Zhou C, et al. Diagnostic value of circulating cell-free mtDNA in patients with suspected thyroid cancer: ND4/ND1 ratio as a new potential plasma marker. Mitochondrion, 2020,55:145–153
Maximo V, Soares P, Seruca R, et al. Microsatellite instability, mitochondrial DNA large deletions, and mitochondrial DNA mutations in gastric carcinoma. Genes Chromosomes Cancer, 2001,32(2):136–143
Ghidini M, Lampis A, Mirchev MB, et al. Immune-Based Therapies and the Role of Microsatellite Instability in Pancreatic Cancer. Genes (Basel), 2020,12(1):33
Jones JB, Song JJ, Hempen PM, et al. Detection of mitochondrial DNA mutations in pancreatic cancer offers a “mass”-ive advantage over detection of nuclear DNA mutations. Cancer Res, 2001,61(4):1299–1304
Okada T, Mizukami Y, Ono Y, et al. Digital PCR-based plasma cell-free DNA mutation analysis for early-stage pancreatic tumor diagnosis and surveillance. J Gastroenterol, 2020,55(12):1183–1193
Warowicka A, Wolun-Cholewa M, Kwasniewska A, et al. Alternations in mitochondrial genome in carcinogenesis of HPV positive cervix. Exp Mol Pathol, 2020,117:104530
Jarviaho T, Hurme-Niiranen A, Soini HK, et al. Novel non-neutral mitochondrial DNA mutations found in childhood acute lymphoblastic leukemia. Clin Genet, 2018,93(2):275–285
Linnartz B, Anglmayer R, Zanssen S. Comprehensive scanning of somatic mitochondrial DNA alterations in acute leukemia developing from myelodysplastic syndromes. Cancer Res, 2004,64(6):1966–1971
Iommarini L, Kurelac I, Capristo M, et al. Different mtDNA mutations modify tumor progression in dependence of the degree of respiratory complex I impairment. Hum Mol Genet, 2014,23(6):1453–1466
Guney AI, Ergec DS, Tavukcu HH, et al. Detection of mitochondrial DNA mutations in nonmuscle invasive bladder cancer. Genet Test Mol Biomarkers, 2012,16(7):672–678
Dmitrenko V, Shostak K, Boyko O, et al. Reduction of the transcription level of the mitochondrial genome in human glioblastoma. Cancer Lett, 2005,218(1):99–107
Jerónimo C, Nomoto S, Caballero OL, et al. Mitochondrial mutations in early stage prostate cancer and bodily fluids. Oncogene, 2001,20(37):5195–5198
Guerra F, Kurelac I, Cormio A, et al. Placing mitochondrial DNA mutations within the progression model of type I endometrial carcinoma. Hum Mol Genet, 2011,20(12):2394–2405
Tsuji A, Akao T, Masuya T, et al. IACS-010759, a potent inhibitor of glycolysis-deficient hypoxic tumor cells, inhibits mitochondrial respiratory complex I through a unique mechanism. J Biol Chem, 2020,295(21):7481–7491
Lomeli N, Di K, Pearre DC, et al. Mitochondrial-associated impairments of temozolomide on neural stem/progenitor cells and hippocampal neurons. Mitochondrion, 2020,52:56–66
Liu Q, Sun Y, Fei Z, et al. Leptin promotes fatty acid oxidation and OXPHOS via the c-Myc/PGC-1 pathway in cancer cells. Acta Biochim Biophys Sin (Shanghai), 2019,51(7):707–714
Ji S, Zheng Z, Liu S, et al. Resveratrol promotes oxidative stress to drive DLC1 mediated cellular senescence in cancer cells. Exp Cell Res, 2018,370(2):292–302
Sitarek P, Synowiec E, Kowalczyk T, et al. An In Vitro Estimation of the Cytotoxicity and Genotoxicity of Root Extract from Leonurus sibiricus L. Overexpressing AtPAP1 against Different Cancer Cell Lines. Molecules, 2018,23(8):2049
Skala E, Synowiec E, Kowalczyk T, et al. Rhaponticum carthamoides Transformed Root Extract Has Potent Anticancer Activity in Human Leukemia and Lung Adenocarcinoma Cell Lines. Oxid Med Cell Longev, 2018,2018:8198652
Liu Z, Ren B, Wang Y, et al. Sesamol Induces Human Hepatocellular Carcinoma Cells Apoptosis by Impairing Mitochondrial Function and Suppressing Autophagy. Sci Rep, 2017,7:45728
Tendi EA, Bosetti F, Dasgupta SF, et al. Ginkgo biloba extracts EGb 761 and bilobalide increase NADH dehydrogenase mRNA level and mitochondrial respiratory control ratio in PC12 cells. Neurochem Res, 2002,27(4):319–323
Sripada L, Singh K, Lipatova AV, et al. hsa-miR-4485 regulates mitochondrial functions and inhibits the tumorigenicity of breast cancer cells. J Mol Med (Berl), 2017,95(6):641–651
Greene J, Segaran A, Lord S. Targeting OXPHOS and the electron transport chain in cancer; Molecular and therapeutic implications. Semin Cancer Biol, 2022,86 (Pt 2):851–859
Sriramkumar S, Sood R, Huntington TD, et al. Platinum-induced mitochondrial OXPHOS contributes to cancer stem cell enrichment in ovarian cancer. J Transl Med, 2022,20(1):246
Jonckheere AI, Smeitink JA, Rodenburg RJ. Mitochondrial ATP synthase: architecture, function and pathology. J Inherit Metab Dis, 2012,35(2):211–225
Shen J, Wan J, Song R, et al. Peripheral blood mitochondrial DNA copy number, length heteroplasmy and breast cancer risk: a replication study. Carcinogenesis, 2015,36(11):1307–1313
Thyagarajan B, Wang R, Barcelo H, et al. Mitochondrial copy number is associated with colorectal cancer risk. Cancer Epidemiol Biomarkers Prev, 2012,21(9):1574–1581
Lee JS, Ko YG, Shin KJ, et al. Mitochondrial DNA 4977bp deletion mutation in peripheral blood reflects atrial remodeling in patients with non-valvular atrial fibrillation. Yonsei Med J, 2015,56(1):53–61
Tan BH, Skipworth RJ, Stephens NA, et al. Frequency of the mitochondrial DNA 4977bp deletion in oesophageal mucosa during the progression of Barrett’s oesophagus. Eur J Cancer, 2009,45(5):736–740
Shaik NA, Lone WG, Khan IA, et al. Detection of somatic mutations and germline polymorphisms in mitochondrial DNA of uterine fibroids patients. Genet Test Mol Biomarkers, 2011,15(7–8):537–541
Gasparre G, Kurelac I, Capristo M, et al. A mutation threshold distinguishes the antitumorigenic effects of the mitochondrial gene MTND1, an oncojanus function. Cancer Res, 2011,71(19):6220–6229
Duan K, Liu ZJ, Hu SQ, et al. Lactic acid induces lactate transport and glycolysis/OXPHOS interconversion in glioblastoma. Biochem Biophys Res Commun, 2018,503(2):888–894
Gasparre G, Iommarini L, Porcelli AM, et al. An inherited mitochondrial DNA disruptive mutation shifts to homoplasmy in oncocytic tumor cells. Hum Mutat, 2009,30(3):391–396
Schöller E, Marks J, Marchand V, et al. Balancing of mitochondrial translation through METTL8-mediated m3C modification of mitochondrial tRNAs. Mol Cell, 2021,81(23):4810–4825
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
The authors declare that there is no conflict of interest with any financial organization or corporation or individual that can inappropriately influence this work.
Additional information
The present study was supported by National Natural Science Foundation of China (No. 82203232), Scientific Instrument Field Project of Shanghai Science and Technology Commission (No. 22142202700) and Shanghai Rising-Star Program (No. 19QB1404700).
Rights and permissions
About this article
Cite this article
Xu, Yc., Su, J., Zhou, Jj. et al. Roles of MT-ND1 in Cancer. CURR MED SCI 43, 869–878 (2023). https://doi.org/10.1007/s11596-023-2771-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11596-023-2771-0