Abstract
Objective
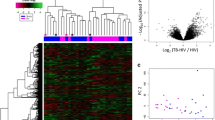
Current commercially available immunological tests cannot be used for discriminating active tuberculosis (TB) from latent TB infection. To evaluate the value of biomarker candidates in the diagnosis of active TB, this study aimed to identify differentially expressed genes in peripheral blood mononuclear cells (PBMCs) between patients with active TB and individuals with latent TB infection by transcriptome sequencing.
Methods
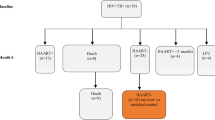
The differentially expressed genes in unstimulated PBMCs and in Mycobacterium tuberculosis (Mtb) antigen-stimulated PBMCs from patients with active TB and individuals with latent TB infection were identified by transcriptome sequencing. Selected candidate genes were evaluated in cohorts consisting of 110 patients with TB, 30 individuals with latent TB infections, and 50 healthy controls by quantitative real-time RT-PCR. Receiver operating characteristic (ROC) curve analysis was performed to calculate the diagnostic value of the biomarker candidates.
Results
Among the differentially expressed genes in PBMCs without Mtb antigen stimulation, interferon-induced protein with tetratricopeptide repeats 3 (IFIT3) had the highest area under curve (AUC) value (0.918, 95% CI: 0.852–0.984, P<0.0001) in discriminating patients with active TB from individuals with latent TB infection, with a sensitivity of 91.86% and a specificity of 84.00%. In Mtb antigen-stimulated PBMCs, orosomucoid 1 (ORM1) had a high AUC value (0.833, 95% CI: 0.752–0.915, P<0.0001), with a sensitivity of 81.94% and a specificity of 70.00%.
Conclusion
IFIT3 and ORM1 might be potential biomarkers for discriminating active TB from latent TB infection.
Similar content being viewed by others
References
WHO. Global Tuberculosis Report 2021. https://www.who.int/publications/i/item/9789240037021, 2021.
Walzl G, McNerney R, du Plessis N, et al. Tuberculosis: advances and challenges in development of new diagnostics and biomarkers. Lancet Infect Dis, 2018,18(7):e199–e210
Boehme CC, Nicol MP, Nabeta P, et al. Feasibility, diagnostic accuracy, and effectiveness of decentralised use of the Xpert MTB/RIF test for diagnosis of tuberculosis and multidrug resistance: a multicentre implementation study. Lancet, 2011,377(9776):1495–1505
Pai M, Denkinger CM, Kik SV, et al. Gamma interferon release assays for detection of Mycobacterium tuberculosis infection. Clin Microbiol Rev, 2014,27(1):3–20
Mazurek GH, Jereb J, Vernon A, et al. Updated guidelines for using Interferon Gamma Release Assays to detect Mycobacterium tuberculosis infection — United States, 2010. MMWR Recomm Rep, 2010,59(RR-5): 1–25
Thillai M, Pollock K, Pareek M, et al. Interferon-gamma release assays for tuberculosis: current and future applications. Expert Rev Respir Med, 2014,8(1):67–78
Singhania A, Wilkinson RJ, Rodrigue M, et al. The value of transcriptomics in advancing knowledge of the immune response and diagnosis in tuberculosis. Nat Immunol, 2018,19(11):1159–1168
Berry MP, Graham CM, McNab FW, et al. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature, 2010,466(7309):973–977
Anderson ST, Kaforou M, Brent AJ, et al. Diagnosis of childhood tuberculosis and host RNA expression in Africa. N Engl J Med, 2014,370(18):1712–1723
Burel JG, Babor M, Pomaznoy M, et al. Host Transcriptomics as a Tool to Identify Diagnostic and Mechanistic Immune Signatures of Tuberculosis. Front Immunol, 2019,10:221
Streitz M, Tesfa L, Yildirim V, et al. Loss of receptor on tuberculin-reactive T-cells marks active pulmonary tuberculosis. PLoS One, 2007,2(8):e735
Portevin D, Moukambi F, Clowes P, et al. Assessment of the novel T-cell activation marker-tuberculosis assay for diagnosis of active tuberculosis in children: a prospective proof-of-concept study. Lancet Infect Dis, 2014,14(10):931–938
Petruccioli E, Petrone L, Vanini V, et al. Assessment of CD27 expression as a tool for active and latent tuberculosis diagnosis. J Infect, 2015,71(5):526–533
Jiang J, Wang X, Wang X, et al. Reduced CD27 expression on antigen-specific CD4+ T cells correlates with persistent active tuberculosis. J Clin Immunol, 2010,30(4): 566–573
Harari A, Rozot V, Bellutti Enders F, et al. Dominant TNF-alpha+ Mycobacterium tuberculosis-specific CD4+ T cell responses discriminate between latent infection and active disease. Nat Med, 2011,17(3):372–376
Yang Q, Xu Q, Chen Q, et al. Discriminating Active Tuberculosis from Latent Tuberculosis Infection by flow cytometric measurement of CD161-expressing T cells. Sci Rep, 2015,5:17918
Riou C, Berkowitz N, Goliath R, et al. Analysis of the Phenotype of Mycobacterium tuberculosis-Specific CD4+ T Cells to Discriminate Latent from Active Tuberculosis in HIV-Uninfected and HIV-Infected Individuals. Front Immunol, 2017,8:968
Lubbers R, Sutherland JS, Goletti D, et al. Complement Component C1q as Serum Biomarker to Detect Active Tuberculosis. Front Immunol, 2018,9:2427
Wang S, Li Y, Shen Y, et al. Screening and identification of a six-cytokine biosignature for detecting TB infection and discriminating active from latent TB. J Transl Med, 2018,16(1):206
Sabir N, Hussain T, Shah SZA, et al. miRNAs in Tuberculosis: New Avenues for Diagnosis and Host-Directed Therapy. Front Microbiol, 2018,9:602
Pollock KM, Whitworth HS, Montamat-Sicotte DJ, et al. T-cell immunophenotyping distinguishes active from latent tuberculosis. J Infect Dis, 2013,208(6):952–968
Wang X, Jiang J, Cao Z, et al. Diagnostic performance of multiplex cytokine and chemokine assay for tuberculosis. Tuberculosis (Edinb), 2012,92(6):513–520
MacLean E, Broger T, Yerlikaya S, et al. A systematic review of biomarkers to detect active tuberculosis. Nat Microbiol, 2019,4(5):748–758
Yerlikaya S, Broger T, MacLean E, et al. A tuberculosis biomarker database: the key to novel TB diagnostics. Int J Infect Dis, 2017,56:253–257
Zhou X, Michal JJ, Zhang L, et al. Interferon induced IFIT family genes in host antiviral defense. Int J Biol Sci, 2013,9(2):200–208
Daugherty MD, Schaller AM, Geballe AP, et al. Evolution-guided functional analyses reveal diverse antiviral specificities encoded by IFIT1 genes in mammals. Elife, 2016,5:e14228
Li D, Swaminathan S. Human IFIT proteins inhibit lytic replication of KSHV: A new feed-forward loop in the innate immune system. PLoS Pathog, 2019,15(2):e1007609
Pichlmair A, Lassnig C, Eberle CA, et al. IFIT1 is an antiviral protein that recognizes 5’-triphosphate RNA. Nat Immunol, 2011,12(7):624–630
Liu XY, Chen W, Wei B, et al. IFN-induced TPR protein IFIT3 potentiates antiviral signaling by bridging MAVS and TBK1. J Immunol, 2011,187(5):2559–2568
Ceciliani F, Lecchi C. The immune functions of alpha1 acid glycoprotein. Curr Protein Pept Sci, 2019,20(6):505–524
Rocanin-Arjo A, Cohen W, Carcaillon L, et al. A meta-analysis of genome-wide association studies identifies ORM1 as a novel gene controlling thrombin generation potential. Blood, 2014, 123(5):777–785
Ganz PA, Shell WE, Tokes ZA. Evaluation of a radioimmunoassay for alpha 1-acid glycoprotein to monitor therapy of cancer patients. J Natl Cancer Inst, 1983,71(1):25–30
Zhou Z, Li Z, Sun Z, et al. S100A9 and ORM1 serve as predictors of therapeutic response and prognostic factors in advanced extranodal NK/T cell lymphoma patients treated with pegaspargase/gemcitabine. Sci Rep, 2016,6:23695
Fassbender K, Fassbender M, Schaberg T, et al. Glycosylation of alpha 1-acid glycoprotein in bacterial lung infections: distinct pattern in tuberculosis. Clin Chem, 1995,41(3):472–473
Santos VS, Goletti D, Kontogianni K, et al. Acute phase proteins and IP-10 as triage tests for the diagnosis of tuberculosis: systematic review and meta-analysis. Clin Microbiol Infect, 2019,25(2):169–177
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The authors declare no conflicts of interest.
Additional information
This work was supported by grants from the Thirteen-Fifth Mega-Scientific Project on “Prevention and Treatment of AIDS, Viral Hepatitis and Other Infectious Diseases” (No. 2017ZX10201301-007-002) and the National Natural Science Foundation of China (No. 82072233).
Rights and permissions
About this article
Cite this article
Yang, Bf., Zhai, F., Yu, S. et al. Evaluation of IFIT3 and ORM1 as Biomarkers for Discriminating Active Tuberculosis from Latent Infection. CURR MED SCI 42, 1201–1212 (2022). https://doi.org/10.1007/s11596-022-2649-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11596-022-2649-6