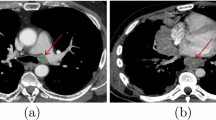
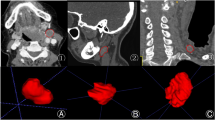
Abstract
Purpose
Identification of lymph nodes (LNs) that are suspicious for metastasis in T2 Magnetic Resonance Imaging (MRI) is critical for assessment of lymphadenopathy. Prior work on LN detection has been limited to specific anatomical regions of the body (pelvis, rectum). Therefore, an approach to universally detect both benign and metastatic nodes in T2 MRI studies of the body is highly desirable.
Methods
We developed a Computer Aided Detection (CAD) pipeline to universally identify LN in T2 MRI. First, we trained various neural networks for detecting LN: Faster RCNN with and without Hard Negative Example Mining (HNEM), FCOS, FoveaBox, VFNet, and Detection Transformer (DETR). Next, we show that VFNet with Adaptive Training Sample Selection (ATSS) outperformed Faster RCNN with HNEM. Finally, we ensembled models that surpassed a 45% mAP threshold.
Results
Experiments on 122 test studies revealed that VFNet achieved a 51.1% mAP and 78.7% recall at 4 false positives (FP) per volume, while the one-stage model ensemble achieved a mAP of 52.3% and sensitivity of 78.7% at 4FP. We found that VFNet and the one-stage model ensemble can be interchangeably used in the CAD pipeline.
Conclusion
Our CAD pipeline universally detected both benign and metastatic nodes in T2 MRI studies, resulting in a sensitivity improvement of \(\sim \)14% over the current LN detection approaches (sensitivity of 78.7% at 4 FP vs. 64.6% at 5 FP per volume).

Similar content being viewed by others
References
Amin MB, Greene FL, Edge SB, Compton CC, Gershenwald JE, Brookland RK, Meyer L, Gress DM, Byrd DR, Winchester DP (2017) The eighth edition ajcc cancer staging manual: continuing to build a bridge from a population-based to a more “personalized’’ approach to cancer staging. CA Cancer J Clin 67(2):93–99
Taupitz M (2007) Imaging of lymph nodes - mri and ct. Springer, pp 321–329
Xingyu Z, Peiyi X, Mengmeng W, Pickhardt Perry J, Wei X, Fei X, Rui Z, Yao X, Junming J (2020) Deep learning based fully automated detection and segmentation of lymph nodes on multiparametric mri for rectal cancer: A multicentre study. EBioMedicine 56:102780
Debats OA, Litjens GJS, Huisman HJ (2019) Lymph node detection in mr lymphography: false positive reduction using multi-view convolutional neural networks. Peer J 7:e8052
Yun L, Qiyue Yu, Gao Y, Zhou Y, Liu G, Dong Q, Ma J, Ding L, Yao H, Zhang Z, Xiao G, An Q, Wang G, Xi J, Yuan W-T, Lian Y, Zhang D, Zhao C-G, Yao Q, Liu W, Zhou X, Liu S, Qingyao W, Wenjian X, Zhang J, Wang D, Sun Z, Gao Y, Zhang X, Jilin H, Zhang M, Wang G, Zheng X, Wang L, Zhao J, Yang S (2018) Identification of metastatic lymph nodes in mr imaging with faster region-based convolutional neural networks. Cancer Res 78 17:5135–5143
Wang S, Zhu Y, Lee S, Elton DC, Shen TC, Tang Y, Peng Y, Lu Z, Summers RM (2022) Global-Local attention network with multi-task uncertainty loss for abnormal lymph node detection in MR images. Med Image Anal 77:102345
Mathai TS, Lee S, Elton DC, Shen TC, Peng Y, Zhiyong L, Summers RM (2021) Detection of lymph nodes in t2 mri using neural network ensembles. In: Lian C, Cao X, Rekik I, Xuanang X, Yan P (eds) Machine Learning in Medical Imaging. Springer International Publishing, Cham
Mathai TS, Lee S, Elton DC, Shen TC, Peng Y, Lu Z, Summers RM (2021) Lymph node detection in t2 mri with transformers. arXiv
Zhang H, Wang Y, Dayoub F, Sünderhauf N (2021) Varifocalnet: An IoU-aware Dense Object Detector, in 2021 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Nashville, TN, USA, 2021 pp. 8510–8519
Carion N, Massa F, Synnaeve G, Usunier N, Kirillov A, Zagoruyko S (2020) End-to-end object detection with transformers. In: Vedaldi A, Bischof H, Brox T, Frahm J-M (eds) Computer vision - ECCV 2020. ECCV 2020. Lecture Notes in Computer Science, vol 12346 Springer International Publishing, Cham
Kong T, Sun F, Liu H, Jiang Y, Li L, Shi J (2020) Foveabox: beyound anchor-based object detection. IEEE Trans Image Process 29:7389–7398
Shaoqing R, Kaiming H, Ross G, Jian S (2015) Faster r-cnn: towards real-time object detection with region proposal networks. In: Cortes C, Lawrence N, Lee D, Sugiyama M, Garnett R (eds) Advances in Neural Information Processing Systems. Curran Associates Inc, Red Hook
Tang Y-B, Yan K, Tang Y-X, Liu J, Xiao J, Summers RM (2019) Uldor: a universal lesion detector for ct scans with pseudo masks and hard negative example mining. In: 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), pp 833–836
Tian Z, Shen C, Chen H, He T (2019) Fcos: fully convolutional one-stage object detection. In: Proceedings of the IEEE/CVF International Conference on Computer Vision (ICCV)
Lin T-Y, Dollár P, Girshick R, He K, Hariharan B, Belongie S (2017) Feature pyramid networks for object detection. In: 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), pp 936–944
Lin T-Y, Goyal P, Girshick R, He K, Dollár P (2020) Focal loss for dense object detection. IEEE Trans Pattern Anal Mach Intell 42(2):318–327
Bodla N, Singh B, Chellappa R, Davis LS (2017) Soft-NMS—improving object detection with one line of code. In: Proceedings of the IEEE International Conference on Computer Vision (ICCV)
Solovyev R, Wang W, Gabruseva T (2021) Weighted boxes fusion: ensembling boxes from different object detection models. Image Vis Comput 107:104117
Peng Y, Lee S, Elton DC, Shen T, Tang Y-X, Chen Q, Wang S, Zhu Y, Summers R, Lu (2020) Automatic recognition of abdominal lymph nodes from clinical text. In: Proceedings of the 3rd clinical natural language processing workshop, pp 101–110, Online, November 2020. Association for Computational Linguistics
Tustison NJ, Avants BB, Cook PA, Zheng Y, Egan A, Yushkevich PA, Gee JC (2010) N4itk: improved n3 bias correction. IEEE Trans Med Imaging 29(6):1310–1320
Kociołek M, Strzelecki M, Obuchowicz R (2020) Does image normalization and intensity resolution impact texture classification? Comput Med Imaging Graph 81:101716
Chen K, Wang J, Pang J, Yuhang Cao Y, Xiong Xiaoxiao L, Sun S, Feng W, Liu Z, Jiarui X, Zhang Z, Cheng D, Zhu C, Cheng T, Zhao Q, Li B, Xin L, Zhu R, Yue W, Dai J, Wang J, Shi J, Ouyang W, Loy Chen C, Lin D (2019) Mmdetection: open mmlab detection toolbox and benchmark
Funding
This work was supported by the Intramural Research Programs of the NIH National Library of Medicine and NIH Clinical Center (project number 1Z01 CL040004). We also thank Jaclyn Burge for the helpful comments and suggestions.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
RMS receives royalties from iCAD, Philips, PingAn, ScanMed, and Translation Holdings. His lab received research support from PingAn. The authors have no additional conflicts of interest to declare.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and the 1964 Helsinki declaration and its later amendments or comparable ethical standards. For this study, informed consent was not required.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Mathai, T.S., Lee, S., Shen, T.C. et al. Universal lymph node detection in T2 MRI using neural networks. Int J CARS 18, 313–318 (2023). https://doi.org/10.1007/s11548-022-02782-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11548-022-02782-1