Abstract
Purpose
Diffeomorphic image registration is essential in many medical imaging applications. Several registration algorithms of such type have been proposed, but primarily for intra-contrast alignment. Currently, efficient inter-modal/contrast diffeomorphic registration, which is vital in numerous applications, remains a challenging task.
Methods
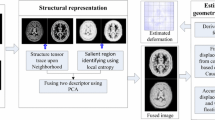
We proposed a novel inter-modal/contrast registration algorithm that leverages Robust PaTch-based cOrrelation Ratio metric to allow inter-modal/contrast image alignment and bandlimited geodesic shooting demonstrated in Fourier-Approximated Lie Algebras (FLASH) algorithm for fast diffeomorphic registration.
Results
The proposed algorithm, named DiffeoRaptor, was validated with three public databases for the tasks of brain and abdominal image registration while comparing the results against three state-of-the-art techniques, including FLASH, NiftyReg, and Symmetric image Normalization (SyN).
Conclusions
Our results demonstrated that DiffeoRaptor offered comparable or better registration performance in terms of registration accuracy. Moreover, DiffeoRaptor produces smoother deformations than SyN in inter-modal and contrast registration. The code for DiffeoRaptor is publicly available at https://github.com/nimamasoumi/DiffeoRaptor.






Similar content being viewed by others
References
Hernandez M, Bossa MN, Olmos S (2009) Registration of anatomical images using paths of diffeomorphisms parameterized with stationary vector field flows. Int J Comput Vis 85:291–306
Tward D, Brown T, Kageyama Y, Patel J, Hou Z, Mori S, Albert M, Troncoso J, Miller M (2020) Diffeomorphic registration with intensity transformation and missing data: application to 3D digital pathology of Alzheimer’s disease. Front Neurosci 14:52
Zhang M, Golland P (2016) Statistical shape analysis: from landmarks to diffeomorphisms. In: Medical image analysis, ISSN: 1361-8415, vol 33, pp 155–158
Blaiotta C, Freund P, Cardoso MJ, Ashburner J (2018) Generative diffeomorphic modelling of large MRI data sets for probabilistic template construction. NeuroImage 166:117–134
Xu Z, Lee CP, Heinrich MP, Modat M, Rueckert D, Ourselin S, Abramson RG, Landman BA (2016) Evaluation of six registration methods for the human abdomen on clinically acquired CT. IEEE Trans Biomed Eng 63(8):1563–1572
Beg M, Miller M, Trouvé A, Younes L (2005) Computing large deformation metric mappings via geodesic flows of diffeomorphisms. Int J Comput Vis 61(2):139–157
Vialard FX, Risser L, Rueckert D, Cotter CJ (2012) Diffeomorphic 3D image registration via geodesic shooting using an efficient adjoint calculation. Int J Comput Vis 97(2):229–241
Zhang M, Fletcher PT (2019) Fast diffeomorphic image registration via Fourier-approximated lie algebras. Int J Comput Vis 127:61–73
Wu J, Tang X (2020) A large deformation diffeomorphic framework for fast brain image registration via parallel computing and optimization. Neuroinformatics 18:251–266
Chen X, Diaz-Pinto A, Ravikumar N, Frangi AF (2021) Deep learning in medical image registration. Prog Biomed Eng 3:012003
Dalca AV, Balakrishnan G, Guttag J, Sabuncu MR (2019) Unsupervised learning of probabilistic diffeomorphic registration for images and surfaces. Med Image Anal 57:226–236
Wang J, Zhang M (2020) Deepflash: an efficient network for learning-based medical image registration. In: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, pp 4444–4452
Hering A, Hansen L, Mok TC, Chung A, Siebert H, Her S, Lange A, Kuckertz S, Heldmann S, Shao W, Vesal S (2021) Learn2Reg: comprehensive multi-task medical image registration challenge, dataset and evaluation in the era of deep learning. arXiv preprint arXiv:2112.04489
Mitra J, Kato Z, Martí R, Oliver A, Lladó X, Sidibé D, Ghose S, Vilanova JC, Comet J, Meriaudeau F (2012) A spline-based non-linear diffeomorphism for multimodal prostate registration. Med Image Anal 16(6):1259–1279
Kutten KS, Charon N, Miller MI, Ratnanather JT, Mateisky J, Baden AD (2017) A large deformation diffeomorphic approach to registration of CLARITY images via mutual information. In: Medical image computing and computer assisted intervention—MICCAI, 2017, vol 10433. Lecture Notes in Computer Science. Springer, Cham
Reaungamornrat S, Silva TD, Uneri A, Vogt S, Kleinszig G, Khanna AJ, Wolinsky JP, Prince JL, Siewerdsen JH (2016) MIND demons: symmetric diffeomorphic deformable registration of MR and CT for image-guided spine surgery. IEEE Trans Med Imaging 35(11):2413–2424
Avants BB, Epstein CL, Grossman M, Gee JC (2008) Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Med Image Anal 12(1):26–41
Vercauteren T, Pennec X, Perchant A, Ayache N (2009) Diffeomorphic demons: efficient non-parametric image registration. Neuroimage 45(1 Suppl):S61-72
Heinrich MP, Jenkinson M, Bhushan M, Matin T, Gleeson FV, Brady M, Schnabel JA (2012) MIND: modality independent neighbourhood descriptor for multi-modal deformable registration. Med Image Anal 16(7):1423–1435
Rivaz H, Chen SJ, Collins DL (2015) Automatic deformable MR-ultrasound registration for image-guided neurosurgery. IEEE Trans Med Imaging 34(2):366–80
Mercier L, Maestro RFD, Petrecca K, Araujo D, Haegelen C, Collins DL (2012) Online database of clinical MR and ultrasound images of brain tumors. Med Phys 39:3253
Masoumi N, Xiao Y, Rivaz H (2019) ARENA: inter-modality affine registration using evolutionary strategy. Int J Comput Assist Radiol Surg 14:441–450
Xiao Y, Fortin M, Unsgård G, Rivaz H, Reinertsen I (2017) REtroSpective evaluation of cerebral tumors (RESECT): a clinical database of pre-operative MRI and intra-operative ultrasound in low-grade glioma surgeries. Med Phys 44:3875–3882
Masoumi N, Belasso CJ, Ahmad MO, Benali H, Xiao Y, Rivaz H (2021) Multimodal 3D ultrasound and CT in image-guided spinal surgery: public database and new registration algorithms. Int J Comput Assist Radiol Surg 16:555–565
Singh N, Hinkle J, Joshi S, Fletcher PT (2013) A vector momenta formulation of diffeomorphisms for improved geodesic regression and atlas construction. In: IEEE 10th international symposium on biomedical imaging, pp 1219–1222
Oliveira FPM, Tavares JMRS (2014) Medical image registration: a review. Comput Methods Biomech Biomed Eng 17(2):73–93
Modat M, Ridgway GR, Taylor ZA, Lehmann M, Barnes J, Hawkes DJ, Fox NC, Ourselin S (2010) Fast free-form deformation using graphics processing units. Comput Methods Program Biomed 98(3):278–284
Miller MI, Trouvé A, Younes L (2006) Geodesic shooting for computational anatomy. J Math Imaging Vis 24:209–228
Joshi SC, Miller MI (2000) Landmark matching via large deformation diffeomorphisms. IEEE Trans Image Process 9(8):1357–1370
Roche A, Malandain G, Pennec X, Ayache N (1998) The correlation ratio as a new similarity measure for multimodal image registration. In: Medical image computing and computer-assisted intervention—MICCAI98, Lecture Notes in Computer Science, vol 1496. Springer, Berlin, Heidelberg
LaMontagne PJ, Benzinger TL, Morris JC, Keefe S, Hornbeck R, Xiong C, Grant E, Hassenstab J, Moulder K, Vlassenko A Raichle ME (2019) OASIS-3: longitudinal neuroimaging, clinical, and cognitive dataset for normal aging and Alzheimer disease. MedRxiv
Clark K, Vendt B, Smith K, Freymann J, Kirby J, Koppel P, Moore S, Phillips S, Maffitt D, Pringle M, Tarbox L, Prior F (2013) The cancer imaging archive (TCIA): maintaining and operating a public information repository. J Digit Imaging 26(6):1045–1057
Avants BB, Tustison N, Song G (2009) Advanced normalization tools (ANTS). Insight j 2(365):1–35
Manjón JV, Coupé P (2016) volBrain: an online MRI brain volumetry system. Front Neuroinform 10:30
Manjón JV, Eskildsen S, Coupé P, Romero J, Collins L, Robles M (2014) NICE: nonlocal intracranial cavity extraction. Int J Biomed Imaging 4191:820205
Grabner G, Janke AL, Budge MM, Smith D, Pruessner J, Collins DL (2006) Symmetric atlasing and model based segmentation: an application to the hippocampus in older adults. Med Image Comput Comput Assist Interv Int Conf Med Image Comput Comput Assist Interv 9:58–66
Rohlfing T (2011) Image similarity and tissue overlaps as surrogates for image registration accuracy: widely used but unreliable. IEEE Trans Med Imaging 31(2):153–163
Acknowledgements
This work was supported in part by the Natural Sciences and Engineering Research Council of Canada (NSERC) and in part by the Regroupment Strategique en Microelectronique du Quebec. Data were provided in part by OASIS-3: Principal Investigators: T. Benzinger, D. Marcus, J. Morris; NIH P50 AG00561, P30 NS09857781, P01 AG026276, P01 AG003991, R01 AG043434, UL1 TR000448, R01 EB009352. AV-45 doses were provided by Avid Radiopharmaceuticals, a wholly owned subsidiary of Eli Lilly.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interest.
Ethical standard
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Declaration of Helsinki and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all participants included in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Masoumi, N., Rivaz, H., Ahmad, M.O. et al. DiffeoRaptor: diffeomorphic inter-modal image registration using RaPTOR. Int J CARS 18, 367–377 (2023). https://doi.org/10.1007/s11548-022-02749-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11548-022-02749-2