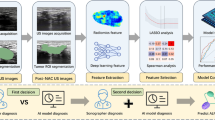
Abstract
Background
The accurate identification and evaluation of lymph nodes by CT images is of great significance for disease diagnosis, treatment, and prognosis.
Purpose
To assess the lymph nodes’ segmentation, size, and station by artificial intelligence (AI) for unenhanced chest CT images and evaluate its value in clinical scenarios.
Material and methods
This retrospective study proposed an end-to-end Lymph Nodes Analysis System (LNAS) consisting of three models: the Lymph Node Segmentation model (LNS), the Mediastinal Organ Segmentation model (MOS), and the Lymph Node Station Registration model (LNR). We selected a healthy chest CT image as the template image and annotated 14 lymph node station masks according to the IASLC to build the lymph node station mapping template. The exact contours and stations of the lymph nodes were annotated by two junior radiologists and reviewed by a senior radiologist. Patients aged 18 and above, who had undergone unenhanced chest CT and had at least one suspicious enlarged mediastinal lymph node in imaging reports, were included. Exclusions were patients who had thoracic surgeries in the past 2 weeks or artifacts on CT images affecting lymph node observation by radiologists. The system was trained on 6725 consecutive chest CTs that from Tianjin Medical University General Hospital, among which 6249 patients had suspicious enlarged mediastinal lymph nodes. A total of 519 consecutive chest CTs from Qilu Hospital of Shandong University (Qingdao) were used for external validation. The gold standard for each CT was determined by two radiologists and reviewed by one senior radiologist.
Results
The patient-level sensitivity of the LNAS system reached of 93.94% and 92.89% in internal and external test dataset, respectively. And the lesion-level sensitivity (recall) reached 89.48% and 85.97% in internal and external test dataset. For man–machine comparison, AI significantly apparently shortened the average reading time (p < 0.001) and had better lesion-level and patient-level sensitivities.
Conclusion
AI improved the sensitivity lymph node segmentation by radiologists with an advantage in reading time.



Similar content being viewed by others
References
Mariën H, Derveaux E, Vanhove K, Adriaensens P, Thomeer M, Mesotten L (2022) Changes in metabolism as a diagnostic tool for lung cancer: systematic review. Metabolites 12:545. https://doi.org/10.3390/metabo12060545
Walker CM, Chung JH, Abbott GF, Little BP, El-Sherief AH, Shepard J-AO, Lanuti M (2012) Mediastinal lymph node staging: from noninvasive to surgical. Am J Roentgenol 199:W54–W64. https://doi.org/10.2214/AJR.11.7446
Rami-Porta R, Asamura H, Travis WD, Rusch VW (2017) ung cancer - major changes in the American Joint Committee on Cancer eighth edition cancer staging manual: The Eighth Edition of the TNM Classification for Lung Cancer. CA Cancer J Clin 67:138–155. https://doi.org/10.3322/caac.21390
Shen D, Wu G, Suk H-I (2017) Deep learning in medical image analysis. Annu Rev Biomed Eng 19:221–248. https://doi.org/10.1146/annurev-bioeng-071516-044442
Cheng PM, Montagnon E, Yamashita R, Pan I, Cadrin-Chênevert A, Perdigón Romero F, Chartrand G, Kadoury S, Tang A (2021) Deep learning: an update for radiologists. RadioGraphics 41:1427–1445. https://doi.org/10.1148/rg.2021200210
Zhou HY, Yu Y, Wang C, Zhang S, Gao Y, Pan J et al (2023) A transformer-based representation-learning model with unified processing of multimodal input for clinical diagnostics. Nat Biomed Eng 7(6):743–755. https://doi.org/10.1038/s41551-023-01045-x
Ahamed KU, Islam M, Uddin A, Akhter A, Paul BK, Yousuf MA, Uddin S, Quinn JMW, Moni MA (2021) A deep learning approach using effective preprocessing techniques to detect COVID-19 from chest CT-scan and X-ray images. Comput Biol Med 139:105014. https://doi.org/10.1016/j.compbiomed.2021.105014
Pereira S, Pinto A, Alves V, Silva CA (2016) Brain tumor segmentation using convolutional neural networks in MRI images. IEEE Trans Med Imaging 35:1240–1251. https://doi.org/10.1109/TMI.2016.2538465
Zhou Y, Xu J, Liu Q, Li C, Liu Z, Wang M, Zheng H, Wang S (2018) A radiomics approach with CNN for shear-wave elastography breast tumor classification. IEEE Trans Biomed Eng 65:1935–1942. https://doi.org/10.1109/TBME.2018.2844188
Hamm CA, Wang CJ, Savic LJ, Ferrante M, Schobert I, Schlachter T, Lin M, Duncan JS, Weinreb JC, Chapiro J, Letzen B (2019) Deep learning for liver tumor diagnosis part I: development of a convolutional neural network classifier for multi-phasic MRI. Eur Radiol 29:3338–3347. https://doi.org/10.1007/s00330-019-06205-9
Valente IRS, Cortez PC, Neto EC, Soares JM, de Albuquerque VHC, Tavares JMRS (2016) Automatic 3D pulmonary nodule detection in CT images: a survey. Comput Methods Programs Biomed 124:91–107. https://doi.org/10.1016/j.cmpb.2015.10.006
Rusch VW, Asamura H, Watanabe H, Giroux DJ, Rami-Porta R, Goldstraw P, Members of IASLC Staging Committee (2009) The IASLC lung cancer staging project: a proposal for a new international lymph node map in the forthcoming seventh edition of the TNM classification for lung cancer. J Thorac Oncol. 4:568–577
El-Sherief AH, Lau CT, Wu CC, Drake RL, Abbott GF, Rice TW (2014) International association for the study of lung cancer (IASLC) lymph node map: radiologic review with CT illustration. Radiographics 34:1680–1691. https://doi.org/10.1148/rg.346130097
Feulner J, Kevin Zhou S, Hammon M, Hornegger J, Comaniciu D (2013) Lymph node detection and segmentation in chest CT data using discriminative learning and a spatial prior. Med Image Anal 17:254–270. https://doi.org/10.1016/j.media.2012.11.001
Liu J, Hoffman J, Zhao J, Yao J, Lu L, Kim L, Turkbey EB, Summers RM (2016) Mediastinal lymph node detection and station mapping on chest CT using spatial priors and random forest: mediastinal lymph node detection and station mapping. Med Phys 43:4362–4374. https://doi.org/10.1118/1.4954009
Wang H, Zhou Z, Li Y, Chen Z, Lu P, Wang W, Liu W, Yu L (2017) Comparison of machine learning methods for classifying mediastinal lymph node metastasis of non-small cell lung cancer from 18F-FDG PET/CT images. EJNMMI Res 7:11. https://doi.org/10.1186/s13550-017-0260-9
Tekchandani H, Verma S, Londhe N (2020) Performance improvement of mediastinal lymph node severity detection using GAN and Inception network. Comput Methods Programs Biomed 194:105478. https://doi.org/10.1016/j.cmpb.2020.105478
Tekchandani H, Verma S, Londhe ND (2020) Mediastinal lymph node malignancy detection in computed tomography images using fully convolutional network. Biocybern Biomed Eng 40:187–199. https://doi.org/10.1016/j.bbe.2019.05.002
Feuerstein M, Glocker B, Kitasaka T, Nakamura Y, Iwano S, Mori K (2012) Mediastinal atlas creation from 3-D chest computed tomography images: application to automated detection and station mapping of lymph nodes. Med Image Anal 16:63–74. https://doi.org/10.1016/j.media.2011.05.005
Barbu A, Suehling M, Xun Xu, Liu D, Zhou SK, Comaniciu D (2012) Automatic detection and segmentation of lymph nodes from CT data. IEEE Trans Med Imaging 31:240–250. https://doi.org/10.1109/TMI.2011.2168234
Liu J, Hoffman J, Zhao J, Yao J, Lu L, Kim L et al (2016) Mediastinal lymph node detection and station mapping on chest CT using spatial priors and random forest. Med Phys 43(7):4362. https://doi.org/10.1118/1.4954009
Iuga AI, Lossau T, Caldeira LL, Rinneburger M, Lennartz S, Grosse Hokamp N et al (2021) Automated mapping and N-Staging of thoracic lymph nodes in contrast-enhanced CT scans of the chest using a fully convolutional neural network. Eur J Radiol 139:109718. https://doi.org/10.1016/j.ejrad.2021.109718
Iuga AI, Carolus H, Hoink AJ, Brosch T, Klinder T, Maintz D et al (2021) Automated detection and segmentation of thoracic lymph nodes from CT using 3D foveal fully convolutional neural networks. BMC Med Imaging 21(1):69. https://doi.org/10.1186/s12880-021-00599-z
Dong M, Hou G, Li S, Li N, Zhang L, Xu K (2021) Preoperatively estimating the malignant potential of mediastinal lymph nodes: a pilot study toward establishing a robust radiomics model based on contrast-enhanced CT imaging. Front Oncol 10:558428. https://doi.org/10.3389/fonc.2020.558428
Spira D, Wecker M, Spira SM, Hetzel J, Spengler W, Sauter A, Horger M (2013) Does volume perfusion computed tomography enable differentiation of metastatic and non-metastatic mediastinal lymph nodes in lung cancer patients? A feasibility study. Cancer Imaging 13:323–331. https://doi.org/10.1102/1470-7330.2013.0033
Krarup MMK, Krokos G, Subesinghe M, Nair A, Fischer BM (2021) Artificial intelligence for the characterization of pulmonary nodules, lung tumors and mediastinal nodes on PET/CT. Semin Nucl Med 51:143–156. https://doi.org/10.1053/j.semnuclmed.2020.09.001
Yoo J, Cheon M, Park YJ, Hyun SH, Zo JI, Um S-W, Won H-H, Lee K-H, Kim B-T, Choi JY (2021) Machine learning-based diagnostic method of pre-therapeutic 18F-FDG PET/CT for evaluating mediastinal lymph nodes in non-small cell lung cancer. Eur Radiol 31:4184–4194. https://doi.org/10.1007/s00330-020-07523-z
Acknowledgements
This study was supported by the National Natural Science Foundation of China (82071907, 82271937), Natural Science Foundation of Tianjin (18JCYBJC25100), Health science and Technology project of Tianjin (MS20022), Wu Jieping Medical Foundation-special Fund for Clinical Research (320.6750.2022-3-5), China International Medical Foundation Sky Imaging Research Fund (Z-2014-07-2003-05), Tianjin Key Medical Discipline (Specialty) Construction Project (TJYXZDXK-001A), Tianjin University of Science and Technology Development Projects Fund (20140115), and Zhangjiakou City Self-financing Project of the 2019 Scientific Research Plan (1921131H).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have nothing to disclose.
Ethical standards
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Cao, Y., Feng, J., Wang, C. et al. LNAS: a clinically applicable deep-learning system for mediastinal enlarged lymph nodes segmentation and station mapping without regard to the pathogenesis using unenhanced CT images. Radiol med 129, 229–238 (2024). https://doi.org/10.1007/s11547-023-01747-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11547-023-01747-x