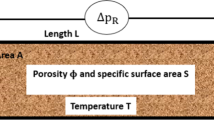
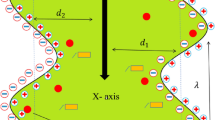
Abstract
The bacterial colony is a powerful experimental platform for broad biological research, and reaction–diffusion models are widely used to study the mechanisms of its formation process. However, there are still some crucial factors that drastically affect the colony growth but are not considered in the current models, such as the non-homogeneously distributed nutrient within the colony and the substantially decreasing expansion rate caused by agar dehydration. In our study, we propose two plausible reaction–diffusion models (the VN and MVN models) based on the above two factors and validate them against experimental data. Both models provide a plausible description of the non-homogeneously distributed nutrient within the colony and outperform the classical Fisher–Kolmogorov equation and its variation in better describing experimental data. Moreover, by accounting for agar dehydration, the MVN model captures how a colony’s expansion slows down and the change of a colony’s height profile over time. Furthermore, we demonstrate the existence of a traveling wave solution for the VN model.









Similar content being viewed by others
References
Basu S, Gerchman Y, Collins CH, Arnold FH, Weiss R (2005) A synthetic multicellular system for programmed pattern formation. Nature 434(7037):1130–1134
Blanchard AE, Lu T (2015) Bacterial social interactions drive the emergence of differential spatial colony structures. BMC Syst Biol 9(1):1–13
Caboussat A, Glowinski R (2009) A numerical method for a non-smooth advection–diffusion problem arising in sand mechanics. Commun Pure Appl Anal 8(1):161
Chacón JM, Möbius W, Harcombe WR (2018) The spatial and metabolic basis of colony size variation. ISME J 12(3):669–680
Chen W, Nie Q, Yi T-M, Chou C-S (2016) Modelling of yeast mating reveals robustness strategies for cell-cell interactions. PLoS Comput Biol 12(7):1004988
Cole JA, Kohler L, Hedhli J, Luthey-Schulten Z (2015) Spatially-resolved metabolic cooperativity within dense bacterial colonies. BMC Syst Biol 9(1):1–17
Dietrich LE, Okegbe C, Price-Whelan A, Sakhtah H, Hunter RC, Newman DK (2013) Bacterial community morphogenesis is intimately linked to the intracellular redox state. J Bacteriol 195(7):1371–1380
Droop MR (1968) Vitamin B12 and marine ecology. IV. The kinetics of uptake, growth and inhibition in Monochrysis lutheri. J Marine Biol Assoc UK 48(3):689–733
Gardner TS, Cantor CR, Collins JJ (2000) Construction of a genetic toggle switch in Escherichia coli. Nature 403(6767):339–342
He C, Bayakhmetov S, Harris D, Kuang Y, Wang X (2020) A predictive reaction-diffusion based model of E. coli colony growth control. IEEE Control Syst Lett 5(6):1952–1957
Huang J, Lu G, Ruan S (2003) Existence of traveling wave solutions in a diffusive predator-prey model. J Math Biol 46(2):132–152
Kawasaki K, Mochizuki A, Matsushita M, Umeda T, Shigesada N (1997) Modeling spatio-temporal patterns generated bybacillus subtilis. J Theor Biol 188(2):177–185
Kondo S, Miura T (2010) Reaction–diffusion model as a framework for understanding biological pattern formation. Science 329(5999):1616–1620
Kreft J-U, Booth G, Wimpenny JW (1998) Bacsim, a simulator for individual-based modelling of bacterial colony growth. Microbiology 144(12):3275–3287
Lacasta A, Cantalapiedra I, Auguet C, Penaranda A, Ramirez-Piscina L (1999) Modeling of spatiotemporal patterns in bacterial colonies. Phys Rev E 59(6):7036
Lagarias JC, Reeds JA, Wright MH, Wright PE (1998) Convergence properties of the Nelder-Mead simplex method in low dimensions. SIAM J Optim 9(1):112–147
Langebrake JB, Dilanji GE, Hagen SJ, De Leenheer P (2014) Traveling waves in response to a diffusing quorum sensing signal in spatially-extended bacterial colonies. J Theor Biol 363:53–61
Leyva JF, Málaga C, Plaza RG (2013) The effects of nutrient chemotaxis on bacterial aggregation patterns with non-linear degenerate cross diffusion. Physica A 392(22):5644–5662
Li B (2018) Multiple invasion speeds in a two-species integro-difference competition model. J Math Biol 76(7):1975–2009
Li X, Gonzalez F, Esteves N, Scharf BE, Chen J (2020) Formation of phage lysis patterns and implications on co-propagation of phages and motile host bacteria. PLoS Comput Biol 16(3):1007236
Luo N, Wang S, You L (2019) Synthetic pattern formation. Biochemistry 58(11):1478–1483
Mansour M (2008) Traveling wave solutions of a nonlinear reaction–diffusion-chemotaxis model for bacterial pattern formation. Appl Math Model 32(2):240–247
Matsushita M, Fujikawa H (1990) Diffusion-limited growth in bacterial colony formation. Physica A 168(1):498–506
Melke P, Sahlin P, Levchenko A, Jönsson H (2010) A cell-based model for quorum sensing in heterogeneous bacterial colonies. PLoS Comput Biol 6(6):1000819
Mimura M, Tsujikawa T (1996) Aggregating pattern dynamics in a chemotaxis model including growth. Physica A 230(3–4):499–543
Mimura M, Sakaguchi H, Matsushita M (2000) Reaction–diffusion modelling of bacterial colony patterns. Physica A 282(1–2):283–303
Nikolopoulou E, Johnson L, Harris D, Nagy J, Stites E, Kuang Y (2018) Tumour-immune dynamics with an immune checkpoint inhibitor. Lett Biomath 5(2):137–159
Ohgiwari M, Matsushita M, Matsuyama T (1992) Morphological changes in growth phenomena of bacterial colony patterns. J Phys Soc Jpn 61(3):816–822
Peñil Cobo M, Libro S, Marechal N, D’Entremont D, Peñil Cobo D, Berkmen M (2018) Visualizing bacterial colony morphologies using time-lapse imaging chamber mocha. J Bacteriol 200(2):00413–17
Peterson JR, Cole JA, Luthey-Schulten Z (2017) Parametric studies of metabolic cooperativity in Escherichia coli colonies: strain and geometric confinement effects. PLoS ONE 12(8):0182570
Potvin-Trottier L, Lord ND, Vinnicombe G, Paulsson J (2016) Synchronous long-term oscillations in a synthetic gene circuit. Nature 538(7626):514–517
Rudge TJ, Steiner PJ, Phillips A, Haseloff J (2012) Computational modeling of synthetic microbial biofilms. ACS Synth Biol 1(8):345–352
Schlüter DK, Ramis-Conde I, Chaplain MA (2015) Multi-scale modelling of the dynamics of cell colonies: insights into cell-adhesion forces and cancer invasion from in silico simulations. J R Soc Interface 12(103):20141080
Sekine R, Shibata T, Ebisuya M (2018) Synthetic mammalian pattern formation driven by differential diffusivity of nodal and lefty. Nat Commun 9(1):1–11
Shapiro JA (1995) The significances of bacterial colony patterns. BioEssays 17(7):597–607
Shin J, Noireaux V (2012) An E. coli cell-free expression toolbox: application to synthetic gene circuits and artificial cells. ACS Synthetic Biol 1(1):29–41
Srinivasan S, Kaplan CN, Mahadevan L (2019) A multiphase theory for spreading microbial swarms and films. Elife 8:42697
Stepien TL, Smith HL (2015) Existence and uniqueness of similarity solutions of a generalized heat equation arising in a model of cell migration. Discrete Contin Dyn Syst 35(7):3203
Stepien TL, Rutter EM, Kuang Y (2018) Traveling waves of a go-or-grow model of glioma growth. SIAM J Appl Math 78(3):1778–1801
Su P-T, Liao C-T, Roan J-R, Wang S-H, Chiou A, Syu W-J (2012) Bacterial colony from two-dimensional division to three-dimensional development. PLoS ONE 7(11):48098
Tyson JJ, Brazhnik PK (2000) On traveling wave solutions of fisher’s equation in two spatial dimensions. SIAM J Appl Math 60(2):371–391
Ward JP, King JR, Koerber AJ, Croft JM, Sockett RE, Williams P (2003) Early development and quorum sensing in bacterial biofilms. J Math Biol 47(1):23–55
Warren MR, Sun H, Yan Y, Cremer J, Li B, Hwa T (2019) Spatiotemporal establishment of dense bacterial colonies growing on hard agar. Elife 8:41093
Wimpenny JW (1979) The growth and form of bacterial colonies. Microbiology 114(2):483–486
Wu F, He C, Fang X, Baez J, Ohnmacht T, Zhang Q, Chen X, Allison KR, Kuang Y, Wang X (2019) A synthetic biology approach to sequential stripe patterning and somitogenesis. bioRxiv 825406
Zhang L, Lander AD, Nie Q (2012) A reaction-diffusion mechanism influences cell lineage progression as a basis for formation, regeneration, and stability of intestinal crypts. BMC Syst Biol 6(1):1–13
Acknowledgements
This work was supported by the National Institutes of Health (NIH) under Grant 5R01GM131405 and the US National Science Foundation (NSF) Rules of Life program DEB-1930728.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
He, C., Han, L., Harris, D.C. et al. Reaction–Diffusion Modeling of E. coli Colony Growth Based on Nutrient Distribution and Agar Dehydration. Bull Math Biol 85, 61 (2023). https://doi.org/10.1007/s11538-023-01163-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11538-023-01163-2