Abstract
Background
Panel-based comprehensive genomic profiling is used in clinical practice worldwide; however, large real-world datasets of patients with advanced gastric cancer are not well known.
Objective
We investigated what differences exist in clinically relevant alterations for molecularly defined or age-stratified subgroups.
Methods
This was a collaborative biomarker study of a real-world dataset from comprehensive genomic profiling testing (Foundation Medicine, Inc.). Hybrid capture was carried out on at least 324 cancer-related genes and select introns from 31 genes frequently rearranged in cancer. Overall, 4634 patients were available for analyses and were stratified by age (≥ 40/< 40 years), microsatellite instability status, tumor mutational burden status (high 10 ≥ /low < 10 Muts/Mb), Epstein–Barr virus status, and select gene alterations. We analyzed the frequency of alterations with a chi-square test with Yate’s correction.
Results
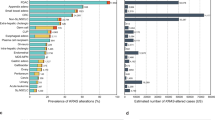
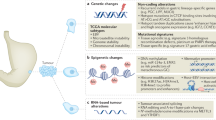
Genes with frequent alterations included TP53 (60.1%), ARID1A (19.6%), CDKN2A (18.2%), KRAS (16.6%), and CDH1 (15.8%). Differences in comprehensive genomic profiling were observed according to molecularly defined or age-stratified subgroups. Druggable genomic alterations were detected in 31.4% of patients; ATM (4.4%), BRAF V600E (0.4%), BRCA1 (1.5%), BRCA2 (2.9%), ERBB2 amplification (9.2%), IDH1 (0.2%), KRAS G12C (0.7%), microsatellite instability-high (4.8%), NTRK1/2/3 fusion (0.13%), PIK3CA mutation (11.4%), and tumor mutational burden-high (9.4%). CDH1 alterations and MET amplification were significantly more frequent in patients aged < 40 years (27.7 and 6.2%) than in those aged ≥ 40 years (14.7 and 4.0%).
Conclusions
Real-world datasets from clinical panel testing revealed the genomic landscape in gastric cancer by subgroup. These findings provide insights for the current therapeutic strategies and future development of treatments in gastric cancer.







Similar content being viewed by others
References
Siegel RL, Miller KD, Fuchs HE, et al. Cancer statistics, 2022. CA Cancer J Clin. 2022;72:7–33.
The Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–9.
Nakamura Y, Kawazoe A, Lordick F, et al. Biomarker-targeted therapies for advanced-stage gastric and gastro-oesophageal junction cancers: an emerging paradigm. Nat Rev Clin Oncol. 2021;18:473–87.
Liu Y, Sethi NS, Hinoue T, et al. Comparative molecular analysis of gastrointestinal adenocarcinomas. Cancer Cell. 2018;33:721–35.
Totoki Y, Saito-Adachi M, Shiraishi Y, et al. Multiancestry genomic and transcriptomic analysis of gastric cancer. Nat Genet. 2023;55:581–94.
Ichikawa H, Nagahashi M, Shimada Y, et al. Actionable gene-based classification toward precision medicine in gastric cancer. Genome Med. 2017;9:93.
Takeda M, Takahama T, Sakai K, et al. Clinical application of the FoundationOne CDx assay to therapeutic decision-making for patients with advanced solid tumors. Oncologist. 2021;26:e588–96.
Inagaki C, Maeda D, Hatake K, et al. Clinical utility of next-generation sequencing-based panel testing under the universal health-care system in Japan: a retrospective analysis at a single university hospital. Cancers (Basel). 2021;13:1121.
Takeda H, Imoto K, Umemoto K, et al. Clinical utility of genomic profiling tests in patients with advanced gastrointestinal cancers. Target Oncol. 2022;17:177–85.
Umemoto K, Yamamoto H, Oikawa R, et al. The molecular landscape of pancreatobiliary cancers for novel targeted therapies from real-world genomic profiling. J Natl Cancer Inst. 2022;114:1279–86.
Kanda M, Terashima M, Kinoshita T, et al. A multi-institutional study to evaluate the feasibility of next-generation sequencing and genomic analysis using formalin-fixed, paraffin-embedded biopsies of gastric cancer. Gastric Cancer. 2023;26:108–15.
Setia N, Wang CX, Lager A, et al. Morphologic and molecular analysis of early-onset gastric cancer. Cancer. 2020;127:103–14.
Zhou Q, Tao F, Qiu L, et al. Somatic alteration characteristics of early-onset gastric cancer. J Oncol. 2022;2022:1498053.
Milbury CA, Creeden J, Yip WK, et al. Clinical and analytical validation of FoundationOne®CDx, a comprehensive genomic profiling assay for solid tumors. PLoS ONE. 2022;17: e0264138.
Woodhouse R, Li M, Hughes J, et al. Clinical and analytical validation of FoundationOne Liquid CDx, a novel 324-Gene cfDNA-based comprehensive genomic profiling assay for cancers of solid tumor origin. PLoS ONE. 2020;15: e0237802.
Chalmers ZR, Connelly CF, Fabrizio D, et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 2017;9:34.
Trabucco SE, Gowen K, Maund SL, et al. A novel next-generation sequencing approach to detecting microsatellite instability and pan-tumor characterization of 1000 microsatellite instability-high cases in 67,000 patient samples. J Mol Diagn. 2019;21:1053–66.
Frampton GM, Fichtenholtz A, Otto GA, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol. 2013;31:1023–31.
Lechner M, Frampton GM, Fenton T, et al. Targeted next-generation sequencing of head and neck squamous cell carcinoma identifies novel genetic alterations in HPV+ and HPV− tumors. Genome Med. 2013;5:49.
Sun JX, He Y, Scanford E, et al. A computational approach to distinguish somatic vs. germline origin of genomic alterations from deep sequencing of cancer specimens without a matched normal. PLoS Comput Biol. 2018;14:e1005965.
Coleman RL, Oza AM, Lorusso D, et al. Rucaparib maintenance treatment for recurrent ovarian carcinoma after response to platinum therapy (ARIEL3): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 2017;390:1949–61.
Huang X, Wojtowicz D, Przytycka TM. Detecting presence of mutational signatures in cancer with confidence. Bioinformatics. 2018;34:330–7.
Franzese N, Fan J, Sharan R, et al. ScalpelSig designs targeted genomic panels from data to detect activity of mutational signatures. J Comput Biol. 2022;29:56–73.
Momozawa Y, Sasai R, Usui Y, et al. Expansion of cancer risk profile for BRCA1 and BRCA2 pathogenic variants. JAMA Oncol. 2022;8:871–8.
Marabelle A, Le DT, Ascierto PA, et al. Efficacy of pembrolizumab in patients with noncolorectal high microsatellite instability/mismatch repair-deficient cancer: results from the phase II KEYNOTE-158 study. J Clin Oncol. 2020;38:1–10.
Graf RP, Fisher V, Creeden J, et al. Real-world validation of TMB and microsatellite instability as predictive biomarkers of immune checkpoint inhibitor effectiveness in advanced gastroesophageal cancer. Cancer Res Commun. 2022;2:1037–48.
Chao J, Fuchs CS, Shitara K, et al. Assessment of pembrolizumab therapy for the treatment of microsatellite instability-high gastric or gastroesophageal junction cancer among patients in the KEYNOTE-059, KEYNOTE-061, and KEYNOTE-062 clinical trials. JAMA Oncol. 2021;7:895–902.
Shitara K, Özgüroglu M, Bang YJ, et al. Molecular determinants of clinical outcomes with pembrolizumab versus paclitaxel in a randomized, open-label, phase III trial in patients with gastroesophageal adenocarcinoma. Ann Oncol. 2021;32:1127–36.
Wu YM, Cieslik M, Lonigro RJ, et al. Inactivation of CDK12 delineates a distinct immunogenic class of advanced prostate cancer. Cell. 2018;173:1770-82.e14.
Sokol ES, Pavlick D, Frampton GM, et al. Pan-cancer analysis of CDK12 loss-of-function alterations and their association with the focal tandem-duplicator phenotype. Oncologist. 2019;12:1526–33.
Liu H, Liu K, Dong Z. Targeting CDK12 for cancer therapy: function, mechanism, and drug discovery. Cancer Res. 2021;81:18–26.
Zhou X, Ji Y, Zhou J. Multiple strategies to develop small molecular KRAS directly bound inhibitors. Molecules. 2023;28:3615.
Saito M, Kono K. Landscape of EBV-positive gastric cancer. Gastric Cancer. 2021;24:983–9.
Gu Y, Zhang P, Wang J, et al. Somatic ARID1A mutation stratifies patients with gastric cancer to PD-1 blockade and adjuvant chemotherapy. Cancer Immunol Immunother. 2023;72:1199–208.
Carneiro F. Familial and hereditary gastric cancer, an overview. Best Pract Res Clin Gastroenterol. 2022;58–59: 101800.
Tanaka Y, Chiwaki F, Kojima S, et al. Multi-omic profiling of peritoneal metastases in gastric cancer identifies molecular subtypes and therapeutic vulnerabilities. Nat Cancer. 2021;2:962–77.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
No external funding was used in the preparation of this article.
Conflict of interest
Kumiko Umemoto has received honoraria from Chugai Pharmaceutical. Naoki Izawa has recived honoraria form Chugai Pharmaceutical. Jay A. Moore and Ethan S. Sokol are employees of Foundation Medicine, Inc. and shareholders in Roche. Yu Sunakawa has recived honoraria and research funding from Chugai Pharmaceutical. Hiroyuki Yamamoto, Hiroyuki Arai, Ritsuko Oikawa, Hiroyuki Takeda, Takuro Mizukami, Yohei Kubota, Ayako Doi, Yoshiki Horie, Takashi Ogura, and Naoki Izawa have no conflicts of interest that are directly relevant to the content of this article.
Ethics approval
Approval for this study, including a waiver of informed consent and a Health Insurance Portability and Accountability Act waiver of authorization, was obtained from the Western Institutional Review Board (protocol #20152817). This study was also approved by the Ethics Committee of St. Marianna University School of Medicine.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Availability of data and material
The sequencing data generated in this study are derived from clinical samples. The data supporting the findings of this study are provided within the paper and its supplementary files. All supplementary information accompanying the different analyses and figures presented in this study are provided in the ESM. Because of Health Insurance Portability and Accountability Act requirements, we are not consented to share individualized patient genomic data that contain potentially identifying or sensitive patient information. Foundation Medicine, Inc. is committed to collaborative data analysis, and we have well-established and widely utilized mechanisms by which investigators can query our core genomic database of > 600,000 de-identified sequenced cancers to obtain aggregated datasets. Requests for collaborative datashares can be made by contacting the corresponding author(s) and completing a study review committee form. Once approved, investigators are required to sign a data transfer agreement. Written proposals are considered at monthly meetings and data transfer agreements expire 18 months from execution of the agreement.
Code availability
Not applicable.
Author contributions
HY designed the study, performed the analyses, and interpreted the data. RO performed the analyses and interpreted the data. YS designed the study and interpreted the data. JM and ES collected the data, performed the analyses, and interpreted the data. All authors contributed to the writing and approved the final manuscript.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yamamoto, H., Arai, H., Oikawa, R. et al. The Molecular Landscape of Gastric Cancers for Novel Targeted Therapies from Real-World Genomic Profiling. Targ Oncol (2024). https://doi.org/10.1007/s11523-024-01052-1
Accepted:
Published:
DOI: https://doi.org/10.1007/s11523-024-01052-1