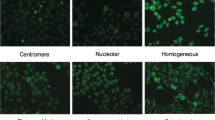
Abstract
In medicine, identifying the indirect immunofluorescence of human epithelial type 2 (HEp-2) cells plays a decisive role in the diagnosis of autoimmune diseases. The manual interpretation of Hep-2 cell images may lead to some limitations, such as subjectivity, inconsistency and low efficiency. Therefore, it is very important to automatically identify HEp-2 images. Inspired by the outstanding performance of neural networks in image classification tasks, we propose a multi-class and multiple-binary classifier (MCMBC) for the classification of HEp-2 cells. MCMBC is an ensemble learner that contains two kinds of sub-classifiers: multi-class (MC) and multiple-binary (MB). The MC sub-classifier adopts a multi-scale convolutional neural network (MSCNN) that increases the efficiency of information transmission between layers. On the basis of classification results of the MC sub-classifier on validation sets, we can find easy-to-confuse class pairs. An easy-to-confuse class pair is two classes that are not easy to be identified from each other. The MB sub-classifiers adopt multiple-binary pre-trained VGG16 networks that are used to deal with these class pairs. The final prediction for a sample possibly belonging to an easy-to-confuse class is decided by the assembled features extracted from the last fully connected layer of MC and the output of MB sub-classifiers. To evaluate the proposed model, experiments were conducted on the ICPR 2014 Task-2 dataset. Experimental results show that MCMBC performs better than the state-of-the-art method (84.68% vs. 83.35% on the criterion of average classification accuracy (ACA) and 82.89% vs. 82.67% on the criterion of mean classification accuracy (MCA)).
Graphical abstract






Similar content being viewed by others
References
Foggia P, Percannella G, Soda P, Vento M (2013) Benchmarking hep-2 cells classification methods. IEEE Trans Med Imaging 32:1878–1889
Li Y, Shen L (2017) A deep residual inception network for HEp-2 cell classification. Deep learning in medical image analysis and multimodal learning for clinical decision support. Springer, Québec City, QC, Canada, pp 12–20
Tonti S, Cataldo SD, Bottino A, Ficarra E (2015) An automated approach to the segmentation of HEp-2 cells for the indirect immunofluorescence ANA test. Comput Med Imaging Graph 40:62–69
Hu C, Zhou R, Zhang S, Qin X, Wu Y, Niu J, Yang Z, He M, Wang C, Lou J (2016) Interpretation of the international consensus on standardized indirect immunofluorescence nomenclature of antinuclear antibody HEp-2 cell patterns and reporting ANA results. Chin J Lab Med 39:804–810
Han XH, Wang J, Xu G, Chen YW (2014) High-order statistics of microtexton for HEp-2 staining pattern classification. IEEE Trans Biomed Eng 61:2223–2234
Ilias T, Dimitris K, George E, Spiros F (2014) HEp-2 cells classification via sparse representation of textural features fused into dissimilarity space. Patt Recognit 47:2367–2378
Lowe DG (2004) Distinctive image features from scale-invariant keypoints. Int J Comput Vis 60:91–110
Nosaka R, Fukui K (2014) HEp-2 cell classification using rotation invariant co-occurrence among local binary patterns. Patt Recognit 47:2428–2436
Thibault G, Angulo J, Meyer F (2014) Advanced statistical matrices for texture characterization: application to cell classification. IEEE Trans Biomed Eng 61:630–637
Wiliem A, Sanderson C, Wong Y, Hobson P, Minchin RF, Lovell BC (2014) Automatic classification of human epithelial type 2 cell indirect immunofluorescence images using cell pyramid matching. Pattern Recognit 47:2315–2324
Xu X, Lin F, Ng C, Leong KP (2015) Automated classification for HEp-2 cells based on linear local distance coding framework. EURASIP J Image Video Process 2015:13
An N, Ding H, Yang J, Au R, Ang TFA (2020) Deep ensemble learning for Alzheimer’s disease classification. J Biomed Inform 105:103411
Harangi B (2018) Skin lesion classification with ensembles of deep convolutional neural networks. J Biomed Inform 86:25–32
Masood A, Sheng B, Li P, Hou X, Wei X, Qin J, Feng D (2018) Computer-assisted decision support system in pulmonary cancer detection and stage classification on ct images. J Biomed Inform 79:117–128
Lecun Y, Bottou L, Bengio Y, Haffuer P (1998) Gradient-based learning applied to document recognition. Proceedings of IEEE 86:2278–2324
Krizhevsky A, Sutskever I, Hinton G (2012) ImageNet classification with deep convolutional neural networks, In: 25th international conference on neural information processing systems, pp 1097–1105
Simonyan K, Zisserman A (2015) Very deep convolutional networks for large-scale image Recognition. 3rd international conference on learning representation. San Diego, CA, USA, pp 1–14
Szegedy C, Liu W, Jia Y, Sermanet P, Reed SE, Anguelov D, Erhan D, Vanhoucke V, Rabinovich A (2015) Going deeper with convolutions. IEEE conference on computer vision and pattern recognition. IEEE Computer Society, Boston, MA, USA, pp 1–9
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. IEEE conference on computer vision and pattern recognition. Las Vegas, NV, USA, pp 770–778
Huang G, Liu Z, Van Der Maaten L, Weinberger KQ (2017) Densely connected convolutional networks, In: 2017 IEEE conference on computer vision and pattern recognition (CVPR), pp 2261–2269
Gao Z, Lei W, Zhou L, Zhang J (2014) HEp-2 cell image classification with deep convolutional neural networks, In: IEEE journal of biomedical and health informatics, IEEE. pp 416–428
Liu J, Xu B, Shen L, Garibaldi J, Qiu G (2017) HEp-2 cell classification based on a deep autoencoding-classification convolutional neural network, In: IEEE 14th international symposium on biomedical imaging, IEEE. pp 1019–1023
Shen L, Jia X, Li Y (2018) Deep cross residual network for HEp-2 cell staining pattern classification. Pattern Recognit 82:68–78
Xie H, He Y, Lei H, Han T, Yu Z, Lei B (2018) Deeply supervised residual network for HEp-2 cell classification. 24th international conference on pattern recognition. IEEE Computer Society, Beijing, China, pp 699–703
Xia X, Yuan Y (2018) Combination of multi-scale convolutional networks and SVM for SAR ATR, In: 2018 2nd IEEE advanced information management, communicates, electronic and automation control conference (IMCEC), pp 66–69
Hobson P, Lovell BC, Percannella G, Saggese A, Vento M, Wiliem A (2016) HEp-2 staining pattern recognition at cell and specimen levels: Datasets, algorithms and results. Patt Recognit Lett 82:12–22
Wiik AS, Høier-Madsen M, Forslid J, Charles P, Meyrowitsch J (2010) Antinuclear antibodies: A contemporary nomenclature using HEp-2 cells. J Autoimmun 35:276–290
Qi X, Zhao G, Chen J, Pietikäinen M (2016) Exploring illumination robust descriptors for human epithelial type 2 cell classification. Pattern Recognit 60:420–429
Deng J, Dong W, Socher R, Li LJ, Li K, Fei-Fei L (2009) ImageNet: A large-scale hierarchical image database. 2009 IEEE computer society conference on computer vision and pattern recognition (CVPR 2009), 20–25 June 2009. Florida, USA, Miami, pp 248–255
Lovell BC, Percannella G, Vento M, Wiliem A (2014) Performance evaluation of indirect immunofluorescence image analysis systems, In: Pattern Recognit, pp 1–25. http://i3a2014.unisa.it/
Robbins H, Monro S (1951) A stochastic approximation method. Ann Math Stat 22:400–407
Maaten LVD, Hinton G (2008) Visualizing data using t-SNE. J Mach Learn Res 9:2579–2605
Zhang L, Zhou W (2016) Fisher-regularized support vector machine. Inform Sci 343(344):79–93
Acknowledgements
We would like to thank anonymous reviewers and Editor Arvind Pathak for their valuable comments and suggestions, which have significantly improved this paper.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supported by the Natural Science Foundation of the Jiangsu Higher Education Institutions of China under Grant No. 19KJA550002, the Six Talent Peak Project of Jiangsu Province of China under Grant No. XYDXX-054, and the Priority Academic Program Development of Jiangsu Higher Education Institutions.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, L., Zhang, MQ. & Lv, X. HEp-2 image classification using a multi-class and multiple-binary classifier. Med Biol Eng Comput 60, 3113–3124 (2022). https://doi.org/10.1007/s11517-022-02646-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11517-022-02646-5