Abstract
Hyperelastic constitutive laws have been commonly used to model the passive behavior of the human skeletal muscle. Despite many efforts, the use of accurate finite element formulations of hyperelastic constitutive laws is still time-consuming for a real-time medical simulation system. The objective of the present study was to develop a deep learning model to predict the hyperelastic constitutive behaviors of the skeletal muscle toward a fast estimation of the muscle tissue stress.
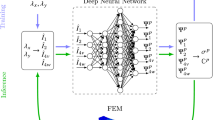
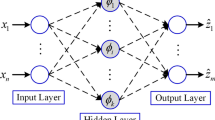
A finite element (FE) model of the right psoas muscle was developed. Neo-Hookean and Mooney-Rivlin laws were used. A tensile test was performed with an applied body force. A learning database was built from this model using an automatic probabilistic generation process. A long-short term memory (LSTM) neural network was implemented to predict the stress evolution of the skeletal muscle tissue. A hyperparameter tuning process was conducted. Root mean square error (RMSE) and associated relative error was quantified to evaluate the precision of the predictive capacity of the developed deep learning model. Pearson correlation coefficients (R) was also computed.
The nodal displacements and the maximal stresses range from 70 to 227 mm and from 2.79 to 5.61 MPa for Neo-Hookean and Monney-Rivlin laws, respectively. Regarding the LSTM predictions, the RMSE ranges from 224.3 ± 3.9 Pa (8%) to 227.5 \(\pm\) 5.7 Pa (4%) for Neo-Hookean and Monney-Rivlin laws, respectively. Pearson correlation coefficients (R) of 0.78 \(\pm\) 0.02 and 0.77 \(\pm\) 0.02 were obtained for Neo-Hookean and Monney-Rivlin laws, respectively.
The present study showed that, for the first time, the use of a deep learning model can reproduce the time-series behaviors of the complex FE formulations for skeletal muscle modeling. In particular, the use of a LSTM neural network leads to a fast and accurate surrogate model for the in silico prediction of the hyperelastic constitutive behaviors of the skeletal muscle. As perspectives, the developed deep learning model will be integrated into a real-time medical simulation of the skeletal muscle for prosthetic socket design and childbirth simulator.
Graphical abstract








Similar content being viewed by others
References
Hill AV (1938) The heat of shortening and the dynamic constants of muscle. Proc R Soc Lond B 126(843):136–195
Tsugorka A, Rios E, Blatter L (1995) Imaging elementary events of calcium release in skeletal muscle cells. Science 269(5231):1723–1726
Wu H, Kanatous SB, Thurmond FA, Gallardo T, Isotani E, Bassel-Duby R, Williams RS (2002) Regulation of mitochondrial biogenesis in skeletal muscle by CaMK. Science 296(5566):349–352
Dao TT, Ho Ba Tho MC (2018) A systematic review of continuum modeling of skeletal muscles: current trends, limitations and recommendations. Appl Bionics Biomech 2018:7631818
Sengupta S, Basak S, Saikia P, Paul S, Tsalavoutis V, Atiah F, Ravi V, Peters A (2020) A review of deep learning with special emphasis on architectures, applications and recent trends. Knowl Based Syst 194:105596
Shrestha A, Mahmood A (2019) Review of deep learning algorithms and architectures. IEEE Access 7:53040–53065. https://doi.org/10.1109/ACCESS.2019.2912200
LeCun Y, Bengio Y, Hinton G (2015) Deep learning. Nature 521:436–444
Kruger N, Janssen P, Kalkan S, Lappe M, Leonardis A, Piater J, Rodriguez-Sanchez AJ, Wiskott L (2013) Deep hierarchies in the primate visual cortex: what can we learn for computer vision?. IEEE Trans Pattern Anal Mach Intell 35(8):1847–1871
LeCun Y, Bottou L, Bengio Y, Haffner P (1998) Gradient-based learning applied to document recognition. Proceedings of IEEE 86(11):2278–2324
Mukherjee P, Zhou M, Lee E, Schicht A, Balagurunathan Y, Napel S, Gillies R, Wong S, Thieme A, Leung A, Gevaert O (2020) A shallow convolutional neural network predicts prognosis of lung cancer patients in multi-institutional computed tomography image datasets. Nat Mach Intell 2:274–282
Kim H, Jung J, Kim J, Cho B, Kwak J, Jang JY, Lee SW, Lee JG, Yoon SM (2020) Abdominal multi-organ auto-segmentation using 3D-patch-based deep convolutional neural network. Sci Rep 10:6204
Hochreiter S, Schmidhuber J (1997) Long short-term memory. Neural Comput 9(8):1735–1780
Dao TT (2019) From deep learning to transfer learning for the prediction of skeletal muscle forces. Med Biol Eng Compu 57(5):1049–1058
Goodfellow I, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S, Courville A, Bengio Y (2014) Generative adversarial networks. Proceedings of the International Conference on Neural Information Processing Systems (NIPS 2014), pp 2672–2680
Kaelbling LP, Littman ML, Moore AW (1996) Reinforcement learning: a survey. J Artif Intell Res 4:237–285
Schmidt J, Marques MRG, Botti S, Marques MAL (2019) Recent advances and applications of machine learning in solid-state materials science. Npj Comput Mater 83. https://doi.org/10.1038/s41524-019-0221-0
Batra R (2021) Accurate machine learning in materials science facilitated by using diverse data sources. Nature 589:524–525
Chen C, Zuo Y, Ye W, Li X, Ong SP (2021) Learning properties of ordered and disordered materials from multi-fidelity data. Nat Comput Sci 1:46–53. https://doi.org/10.1038/s43588-020-00002-x
Liang L, Liu M, Sun W (2017) A deep learning approach to estimate chemically-treated collagenous tissue nonlinear anisotropic stress-strain responses from microscopy images. Acta Biomater 63:227–235
Halilaj E, Rajagopal A, Fiterau M, Hicks JL, Hastie TJ, Delp SL (2018) Machine learning in human movement biomechanics: best practices, common pitfalls, and new opportunities. J Biomech 81:1–11
Liang L, Mao W, Sun W (2020) A feasibility study of deep learning for predicting hemodynamics of human thoracic aorta. J Biomech 99:109544. https://doi.org/10.1016/j.jbiomech.2019.109544
Bustamante-Orellana C, Guachi R, Guachi-Guachi L, Novelli S, Campana F, Bini F, Marinozzi F (2020) Biomechanics of soft tissues: the role of the mathematical model on material behavior. In: Botto-Tobar M, León-Acurio J, Díaz Cadena A, Montiel Díaz P (eds) Advances in Emerging Trends and Technologies. ICAETT 2019. Advances in intelligent systems and computing, vol 1066. Springer. https://doi.org/10.1007/978-3-030-32022-5_29
Affagard J, Bensamoun SF, Feissel P (2014) Development of an inverse approach for the characterization of in vivo mechanical properties of the lower limb muscles. J Biomech Eng 136(11):111012. https://doi.org/10.1115/1.4028490
Avril S, Bouten L, Dubuis L, Drapier S, Pouget J (2010) Mixed experimental and numerical approach for characterizing the biomechanical response of the human leg under elastic compression. J Biomech Eng 132(3):031006
Lapeer R, Gerikhanov Z, Sadulaev SM, Audinis V, Rowland R, Crozier K, Morris E (2019) A computer-based simulation of childbirth using the partial Dirichlet-Neumann contact method with total Lagrangian explicit dynamics on the GPU. Biomech Model Mechanobiol 18:681–700. https://doi.org/10.1007/s10237-018-01109-x
Linder-Ganz E, Shabshin N, Itzchak Y, Gefen A (2007) Assessment of mechanical conditions in sub-dermal tissues during sitting: a combined experimental-MRI and finite element approach. Biomech Model Mechanobiol 40(7):1443–1454
Wu T, Hung A, Mithraratne K (2014) Generating facial expressions using an anatomically accurate biomechanical model. IEEE Trans Visual Comput Graphics 20(11):1519–1529
Lee W, Won BH, Cho SW (2016) Finite element modeling for predicting the contact pressure between a foam mattress and the human body in a supine position. Comput Methods Biomech Biomed Eng 20(1):104–117
Noakes KF, Pullan AJ, Bissett IP, Cheng LK (2008) Subject specific finite elasticity simulations of the pelvic floor. J Biomech 41(14):3060–3065
Sadler Z, Scott J, Drost J, Chen S, Roccabianca S, Bush TR (2018) Initial estimation of the in vivo material properties of the seated human buttocks and thighs. Int J Non-Linear Mech 107:77–85
Silva E, Parente M, Brandão S, Mascarenhas T, Natal RJ (2019) Characterizing the biomechanical properties of the pubovisceralis muscle using a genetic algorithm and the finite element method. J Biomech Eng 141(1)
CoxSL, Mithraratne K, Smith NP (2007) An anatomically based finite element model of the lower limbs in the seated posture. 29th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, pp 6326–6329. https://doi.org/10.1109/IEMBS.2007.4353802.
Dao TT, Marin F, Pouletaut P, Aufaure P, Charleux F, Ho Ba Tho MC (2012) Estimation of accuracy of patient specific musculoskeletal modeling: case study on a post polio residual paralysis subject. Comput Method Biomech Biomed Eng 15(7):745–751
Dao TT (2017) Advanced computational workflow for the multi-scale modeling of the bone metabolic processes. Med Biol Eng Comput 55(6):923–933
Dao TT, Pouletaut P, Charleux F, Lazáry Á, Eltes P, Varga PP, Ho Ba Tho MC (2015) Multimodal medical imaging (CT and dynamic MRI) data and computer-graphics multi-physical model for the estimation of patient specific lumbar spine muscle forces. Data Knowl Eng 96–97:3–18
Meister F, Passerini T, Mihalef V, Tuysuzoglu A, Maier A, Mansi T (2020) Deep learning acceleration of Total Lagrangian explicit dynamics for soft tissue mechanics. Comput Methods Appl Mech Eng 358:112628
ter Horst R, van Weert H, Loonen T, Bergé S, Vinayahalingam S, Baan F, Maal T, de Jong G, Xi T (2021) Three-dimensional virtual planning in mandibular advancement surgery: soft tissue prediction based on deep learning. J Craniomaxillofac Surg. https://doi.org/10.1016/j.jcms.2021.04.001
Medvedev AV, Agoureeva GI, Murro AM (2019) A Long short-term memory neural network for the detection of epileptiform spikes and high frequency oscillations. Sci Rep 9:19374. https://doi.org/10.1038/s41598-019-55861-w
Kumar S, Sharma A, Tsunoda T (2019) Brain wave classification using long short-term memory network based OPTICAL predictor. Sci Rep 9:9153. https://doi.org/10.1038/s41598-019-45605-1
Qiu J, Wang B, Zhou C (2020) Forecasting stock prices with long-short term memory neural network based on attention mechanism. PLoS ONE 15(1):e0227222
Funding
This present study was funded by the Métropole Européenne de Lille (MEL) and the I-SITE ULNE (Université Lille Nord Europe).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Ballit, A., Dao, TT. Recurrent neural network to predict hyperelastic constitutive behaviors of the skeletal muscle. Med Biol Eng Comput 60, 1177–1185 (2022). https://doi.org/10.1007/s11517-022-02541-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11517-022-02541-z