Abstract
Machine learning techniques have been utilized on gene expression profiling for cancer diagnosis. However, the gene expression data suffer from the curse of high dimensionality. Different kinds of feature reduction methods have been proposed to decrease the features for specific cancer diagnosis. However, with the difficulty of obtaining the samples of a particular tumor, the lack of training samples may lead to the overfitting problem. In addition, the feature reduction model on a specific tumor may lead to the problem that the model is not scalable and cannot be generalized to new cancer types. To handle these problems, this paper proposes an unsupervised feature learning method to reduce the data dimensionality of gene expression data. This method amplifies the training samples of feature learning by utilizing the unlabeled samples from different sources. Two heuristic rules are devised to check if the unlabeled samples could be used for amplifying the training set. The amplified training set is used to train the feature learning model based on sparse autoencoder. Since the method leverages the knowledge among the expression data from different sources, it improves the generalization of unsupervised feature learning and further boosts the cancer diagnosis performance. A series of experiments are carried out on the gene expression datasets from TCGA and other sources. Experimental results prove that our method improves the generalization of cancer diagnosis when unlabeled data are used for latent feature learning.
Graphical abstract
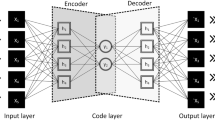
The flowchart of our proposed feature learning method















Similar content being viewed by others
Notes
The Cancer Genome Atlas (TCGA) Research Network. Available: https://www.cancer.gov/ tcga.
References
Salem H, Attiya G, El-Fishawy N (2017) Classification of human cancer diseases by gene expression profiles. Appl Soft Comput 50:124–134
Liu JX, Xu Y, Zheng C-H, Kong H, Lai Z-H (2015) RPCA-based tumor classification using gene expression data. IEEE/ACM Trans Comput Biol Bioinf 12(4):964–970
Mignone P, Pio G, Džeroski S, Ceci M (2020) Multi-task learning for the simultaneous reconstruction of the human and mouse gene regulatory networks. Scientific reports 10:22295
Erola P, Björkegren JLM, Michoel T (2020) Model-based clustering of multi-tissue gene expression data. Bioinformatics 36(6):1807–1813
Bao W, Yuan CA, Zhang Y, Han K, Nandi AK, Honig B, Huang D (2017) Mutli-features prediction of protein translational modification sites. IEEE/ACM Trans Comput Biol Bioinforma 15(5):1453–1460
Bao W, Dong W, Chen Y (2017) Classification of protein structure classes on flexible neutral tree. IEEE/ACM trans comput biol bioinforma 14(5):1122–1133
Yuan F, Lu L, Zou Q (2020) Analysis of gene expression profiles of lung cancer subtypes with machine learning algorithms. BBA-Mol Basis of Dis 866:165822
Khorshed TA (2020) Deep learning for multi-tissue cancer classification of gene expressions (GeneXNet). IEEE Access 8:90615–90629
Tirumala SS, Narayanan A (2016) Attribute selection and classification of prostate cancer gene expression data using artificial neural networks. Pacific-asia Conference on Knowledge Discovery & Data Mining 2016:206–234
Khorshed T, Moustafa MN, Rafea A (2020) Multi-tissue cancer classification of gene expressions using deep learning. IEEE Sixth International Conference on Big Data Computing Service and Applications (BigDataService) 2020:128–135
Abdulla M, Khasawneh MT (2020) G-Forest: an ensemble method for cost-sensitive feature selection in gene expression microarrays. Artif Intell Med 108:101941
Hira ZM, Gillies DF (2015) A review of feature selection and feature extraction methods applied on microarray data. Adv Bioinforma 2015:198363
Hall MA, Smith LA (1998) Practical feature subset selection for machine learning, 21st Australasian Computer Science Conference (ACSC ’98), 1998, pp. 1–11.
Hall MA (2000) Correlation-based feature selection for discrete and numeric class machine learning, 17th International Conference on Machine Learning (ICML’00), 2000, pp. 359–366.
Peng H, Long F, Ding C (2005) Feature selection based on mutual information criteria of max-dependency, max-relevance, and min-redundancy. IEEE Trans Pattern Anal Mach Intell 27(8):1226–1238
Perez M, Marwala T (2012) Microarray data feature selection using hybrid genetic algorithm simulated annealing, IEEE 27th Convention of Electrical and Electronics Engineers in Israel (IEEEI ’12), 2012, pp. 1–5.
Tang EK, Suganthan PN, Yao X (2006) Gene selection algorithms for microarray data based on least squares support vector machine. BMC Bioinformatics 7(95):1–16
Dashtban M, Balafar M (2017) Gene selection for microarray cancer classification using a new evolutionary method employing artificial intelligence concepts. Genomics 109(2):91–107
Lu H, Chen J, Yan K, Jin Q, Xue Y, Gao Z (2017) A hybrid feature selection algorithm for gene expression data classification. Neurocomputing 256:56–62
Jonnalagadda S, Srinivasan R (2008) Principal components analysis based methodology to identify differentially expressed genes in time-course microarray data. BMC Bioinformatics 9:267
Sevakula RK, Singh V, Verma NK, Kumar C, Cui Y (2019) Transfer learning for molecular cancer classification using deep neural networks. IEEE/ACM Trans Comput Biol Bioinf 16(6):2089–2100
Fakoor R, Ladhak F, Nazi A, Huber M (2013) Using deep learning to enhance cancer diagnosis and classification, the 30th International Conference on Machine Learning (ICML 2013), 2013, pp. 1–8.
Liao Q, Ding Y, Jiang ZL, Wang X, Zhang C, Zhang Q (2019) Multi-task deep convolutional neural network for cancer diagnosis. Neurocomputing 348:66–73
Liu Z, Wang R, Zhang W, Tang D (2021) An unsupervised feature learning method for enhancing the generalization of cancer diagnosis. 13th International Conference on Machine Learning and Computing, 2021, pp. 252–257.
Sun L, Zhang X, Qian Y, Xu J, Zhang S (2019) Feature selection using neighborhood entropy-based uncertainty measures for gene expression data classification. Inf Sci 502:18–41
Almugren N, Alshamlan H (2019) A survey on hybrid feature selection methods in microarray gene expression data for cancer classification. IEEE Access 7:78533–78548
Potharaju SP, Sreedevi M (2019) Distributed feature selection (DFS) strategy for microarray gene expression data to improve the classification performance. Clin Epidemiol Glob Health 7:171–176
Wahid A, Khan DM, Iqbal N, Khan SA, Ali A, Khan M, Khan Z (2020) Feature selection and classification for gene expression data using novel correlation based overlapping score method via Chou’s 5-Steps rule. Chemom Intell Lab Syst 199:103958
Uzma, Al-Obeidat F, Tubaishat A, Shah B, Halim Z (2020) Gene encoder: a feature selection technique through unsupervised deep learning-based clustering for large gene expression data, Neural Computing and Applications 2020: 1–23 (published online).
Manosij G, Sukdev A, Kanti GK, Aritra S, Shemim B, Ram S (2019) Genetic algorithm based cancerous gene identification from microarray data using ensemble of filter methods. Med Biol Eng Compu 57:159–176
Nikulin V, McLachlan GJ (2009) Penalized principal component analysis of microarray data. Computational Intelligence Methods for Bioinformatics and Biostatistics 2009:82–96
Huynh PH, Nguyen VH, Do TN (2018) A coupling support vector machines with the feature learning of deep convolutional neural networks for classifying microarray gene expression data. In book: Modern Approaches for Intelligent Information and Database Systems 2018:233–243
Danaee P, Ghaeini R, Hendrix DA (2016) A deep learning approach for cancer detection and relevant gene identification. Pac Symp Biocomput Pac Symp Biocomput 22:219–229
Zhu Z, Ong YS, Dash M (2007) Markov blanket-embedded genetic algorithm for gene selection. Pattern Recogn 49(11):3236–3248
Hess KR (2006) Pharmacogenomic predictor of sensitivity to preoperative chemotherapy with paclitaxel and fluorouracil, doxorubicin, and cyclophosphamide in breast cancer. J Clin Oncol 24(26):4236–4244
Chin K, DeVries S, Fridlyand J, Spellman PT, Roydasgupta R, Kuo W, Lapuk A, Neve RM, Qian Z, Ryder T et al (2006) Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell 10(6):529–541
Telikani A, Gandomi AH (2009) Cost-sensitive stacked auto-encoders for intrusion detection in the Internet of Things. Internet of Things 14:100122
Acknowledgements
We thank the anonymous reviewers for their constructive comments. An earlier version of this paper [24] was presented at the International Conference on the 13th International Conference on Machine Learning and Computing.
Funding
This work is supported by the Key Research Platforms and Projects of Colleges and Universities in Guangdong Province [Grant Nos. 2020ZDZX3060 and 2019KZDZX1020], National Natural Science Foundation of China [Grant No. 61501128], financial support from China Scholarship Council, and Natural Science Foundation of Guangdong Province [Grant No. 2017A030313345].
Key Laboratory of Microbial Resources and Drug Development in Guizhou Province, 2020ZDZX3060, Zhen Liu, National Natural Science Foundation of China, 61501128, Zhen Liu, Natural Science Foundation of Guangdong Province, 2017A030313345, Ruoyu Wang
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Liu, Z., Wang, R. & Zhang, W. Improving the generalization of unsupervised feature learning by using data from different sources on gene expression data for cancer diagnosis. Med Biol Eng Comput 60, 1055–1073 (2022). https://doi.org/10.1007/s11517-022-02522-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11517-022-02522-2