Abstract
The Golgi apparatus and membrane tubules derived from this organelle play essential roles in membrane trafficking in eukaryotic cells. High-resolution live cell imaging is one highly suitable method for studying the molecular mechanisms of dynamics of organelles during membrane trafficking events. Due to the complex morphological changes and dynamic movements of the Golgi apparatus and associated membrane tubules during membrane trafficking, it is challenging to accurately quantify them. In this study, a semi-automated 2D tracking system, 2D-GolgiTrack, has been established for quantifying morphological changes and movements of Golgi elements, specifically encompassing the Golgi apparatus and its associated tubules, the fission and fusion of Golgi tubules, and the kinetics of formation of Golgi tubules and redistribution of the Golgi-associated protein Rab6A to the endoplasmic reticulum. The Golgi apparatus and associated tubules are segmented by a combination of Otsu’s method and adaptive local normalization thresholding. Curvilinear skeletons and tips of skeletons of segmented tubules are used for calculating tubule length by the Geodesic method. The k-nearest neighbor is applied to search the possible candidate objects in the next frame and link the correct objects of adjacent frames by a tracking algorithm to calculate changes in morphological features of each Golgi object or tubule, e.g., number, length, shape, branch point and position, and fission or fusion events. Tracked objects are classified into morphological subtypes, and the Track-Map function of morphological evolution visualizes events of fission and fusion. Our 2D-GolgiTrack not only provides tracking results with 95% accuracy, but also maps morphological evolution for fast visual interpretation of the fission and fusion events. Our tracking system is able to characterize key morphological and dynamic features of the Golgi apparatus and associated tubules, enabling biologists to gain a greater understanding of the molecular mechanisms of membrane traffic involving this essential organelle.

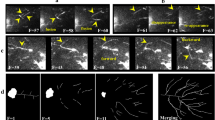
Overview of the semi-automated 2D tracking system. There are two main parts to the system, namely detection and tracking. The workflow process requires a raw sequence of images (a), which is filtered by the Gaussian filter method (c), and threshold intensity (b) to segment elements of Golgi cisternae (d) and tubules (e). Post-processing outputs are binary images of the cisternae area and tubule skeletons. The tubules are classified into three lengths, namely short, medium, and long tubules (f). Outputs of segmentation are calculated as morphological features (g). The tracking processing starts by loading the segmented outputs (h) and key-inputs of direction reference (i; (DR)) and interval setting of the start ((S)) and end ((E)) frame numbers (j). A tubule of interest is selected by the user (k; (GTinterest, S) as the tubule input ((GTIN)) at the current frame ((i = S)). The tracking algorithm tracks and links the correct tubules at each subsequent frame ((i = i + 1)). The locations of tubule tips are determined for detecting tubule branches using the (DR) to identify the direction of tubule growth (l: (1); (GTtipBr, i); Golgi cisternae: white area; Golgi tubule: white skeleton; tubule tips: green dots; branched tubules: two branches due to the (DR) of growth of the simulated tubule moving from left-to-right away from the Golgi cisternae location). According to the position of the (GTIN), five candidates ((GTcandidates, i)) are searched using the k-nearest neighbor method (l: (2)). Matching of tubules between the (GTIN) and those (GTcandidates, i) uses the bounding box technique to check the amount of tubule-overlap based on the tracking conditions (l: (3)). If there is tubule-overlap, the system collects that tubule as the final output ((GTOUT, i)). By contrast, shape (see the Extent feature in Table reftab:1) and distance features are used to generate the tracked output, which has a priority of a minimum of both of these features ((MinDIST, EXTENT)); otherwise, it is from the minimum of the distance ((MinDIST)). Once a loop of the interval track to the last frame is finished ((i = E + 1)), a Track-Map is generated allowing visualization of the morphological pattern of tubule formation and movement, including identification of fission and fusion events (m). Dynamic features are calculated (n). Related outputs are saved, and all features obtained from the detection and tracking processing are exported as MS Excel files (o).













Similar content being viewed by others
Abbreviations
- GC:
-
Golgi cisternae
- GT:
-
Golgi-derived tubules
- ER:
-
endoplasmic reticulum
- COPI:
-
coat protein complex I
- ARF1:
-
ADP-ribosylation factor-1
- BFA:
-
Brefeldin A
- TGN:
-
trans-Golgi network
- YFP:
-
yellow fluorescent protein
- DR:
-
direction reference
- ALNT:
-
adaptive local normalization thresholding
- k NN:
-
k-nearest neighbor
- MSD:
-
mean-square displacement
References
Chen X, Williams J A (2003) Encyclopedia of gastroenterology. In: Johnson L R (ed) Exocytosis. Elsevier, Massachusetts, pp 777–779
Patterson G H, Hirschberg K, Polishchuk R S, Gerlich D, Phair R D, Lippincott-Schwartz J (2008) Transport through the Golgi apparatus by rapid partitioning within a two-phase membrane system. Cell 133(6):1055–1067. https://doi.org/10.1016/j.cell.2008.04.044
Tenorio M J, Luchsinger C, Mardones G A (2015) Protein kinase A activity is necessary for fission and fusion of Golgi to Endoplasmic Reticulum retrograde tubules. PLoS One 10(8):e0135260. https://doi.org/10.1371/journal.pone.0135260
Valente C, Polishchuk R, De Matteis M A (2010) Rab6 Myosin II at the cutting edge of membrane fission. Nat Cell Biol 12(7):635–638. https://doi.org/10.1038/ncb0710-635
Bottanelli F, Kilian N, Ernst A M, Rivera-Molina F, Schroeder L K, Kromann E B, Lessard M D, Erdmann R S, Schepartz A, Baddeley D, Bewersdorf J, Toomre D, Rothman J E (2017) A novel physiological role for ARF1 in the formation of bidirectional tubules from the Golgi. Mol Biol Cell 28 (12):1676–1687. https://doi.org/10.1091/mbc.e16-12-0863
Heffernan L F, Simpson J C (2014) The trials and tubule-ations of Rab6 involvement in golgi-to-ER retrograde transport. Biochem Soc Trans 42(5):1453–1459. https://doi.org/10.1042/BST20140178
Chen L, Liu T, Tran A, Lu X, Tomilov A A, Davies V, Cortopassi G, Chiamvimonvat N, Bers D M, Votruba M, Knowlton A A (2012) OPA1 Mutation and late-onset cardiomyopathy: mitochondrial dysfunction and mtDNA instability. J Am Heart Assoc 1(5):e003012. https://doi.org/10.1161/JAHA.112.003012
Twig G, Elorza A, Molina A, Mohamed H, Wikstrom J D, Walzer G, Stiles L, Haigh S E, Katz S, Las G, Alroy J, Wu M, Py B F, Yuan J, Deeney J T, Corkey B E, Shirihai O S (2008) Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J 27(2):433–46. https://doi.org/10.1038/sj.emboj.7601963
Wang X, Su B, Lee H, Li X, Perry G, Smith M A, Zhu X (2009) Impaired balance of mitochondrial fission and fusion in Alzheimer’s Disease. J Neurosci 29(28):9090–103. https://doi.org/10.1523/JNEUROSCI.1357-09.2009
Knott A B, Perkins G, Schwarzenbacher R, Bossy-Wetzel E (2008) Mitochondrial fragmentation in neurodegeneration. Nat Rev Neurosci 9(7):505–18. https://doi.org/10.1038/nrn2417
Lee Y J, Nagano Y, Taylor J P, Lim K L, Yao T P (2010) Disease-causing mutations in Parkin impair mitochondrial ubiquitination, aggregation, an HDAC6-dependent mitophagy. J Cell Biol 189(4):671–9. https://doi.org/10.1083/jcb.201001039
Bexiga M G, Simpson J C (2013) Human diseases associated with form and function of the Golgi complex. Int J Mol Sci 14(9):18670–81. https://doi.org/10.3390/ijms140918670
Pepperkok R, Simpson J C, Rietdorf J, Cetin C, Liebel U, Terjung S, Zimmermann T (2005) Imaging platforms for measurement of membrane trafficking. Methods Enzymol 404:8–18. https://doi.org/10.1016/S0076-6879(05)04002-4
Neumann B, Walter T, Hériché J K, Bulkescher J, Erfle H, Conrad C, Rogers P, Poser I, Held M, Liebel U, Cetin C, Sieckmann F, Pau G, Kabbe R, Wünsche A, Satagopam V, Schmitz M H, Chapuis C, Gerlich D W, Schneider R, Eils R, Huber W, Peters J M, Hyman A A, Durbin R, Pepperkok R, Ellenberg J (2010) Phenotypic profiling of the human genome by time-lapse microscopy reveals cell division genes. Nature 464(7289):721–727. https://doi.org/10.1038/nature08869
Chenouard N, Smal I, Chaumont F, Maška M, Sbalzarini I F, Gong Y, Cardinale J, Carthel C, Coraluppi S, Winter M, Cohen A R, Godinez W J, Rohr K, Kalaidzidis Y, Liang L, Duncan J, Shen H, Xu Y, Magnusson K E G, Jaldén J, Blau H M, Paul-Gilloteaux P, Roudot P, Kervrann C, Waharte F, Tinevez J -Y, Shorte S L, Willemse J, Celler K, Wezel G P, Dan H -W, Tsai Y -S, Solórzano C, Olivo-Marin J -C, Meijering E (2014) Objective comparison of particle tracking methods. Nat Methods 11(3):281–289. https://doi.org/10.1038/nmeth.2808
Meijering E, Dzyubachyk O, Smal I (2012) Methods for cell and particle tracking. Methods Enzymol 504:183–200. https://doi.org/10.1016/B978-0-12-391857-4.00009-4
Wang S -Q, Fu C -L, Zhang Y, Chen Q, Long M (2010) Dynamics of morphological changes for mitochondrial fission and fusion. Sci China Phys Mech 53(4):680–689. https://doi.org/10.1007/s11433-010-0163-5
Westrate L M, Drocco J A, Martin K R, Hlavacek W S, MacKeigan J P (2014) Mitochondrial morphological features are associated with fission and fusion events. PLoS One 9(4):e95265. https://doi.org/10.1371/journal.pone.0095265
Wait E, Winter M, Bjornsson C, Kokovay E, Wang Y, Goderie S, Temple S, Cohen A R (2014) Visualization and correction of automated segmentation, tracking and lineaging from 5-D stem cell image sequences. BMC Bioinform 15:328. https://doi.org/10.1186/1471-2105-15-328
Matov A, Applegate K, Kumar P, Thoma C, Krek W, Danuser G, Wittmann T (2010) Analysis of microtubule dynamic instability using a plus-end growth marker. Nat Methods 7(9):761–768. https://doi.org/10.1038/nmeth.1493
Applegate KT, Besson S, Matov A, Bagonis MH, Jaqaman K, Danuser G (2011) plusTipTracker: Quantitative image analysis software for the measurement of microtubule dynamics. J Struct Biol 176(2):168–184. https://doi.org/10.1016/j.jsb.2011.07.009
Saban M, Altinok A, Peck A, Kenney C, Feinstein S, Wilson L, Rose K, Manjunath BS (2006) Automated tracking and modeling of microtubule dynamics. In: Proceedings of the IEEE international symposium on biomedical imaging: nano to macro. IEEE, pp 1032–1035. https://doi.org/10.1109/ISBI.2006.1625097
Nussbaum-Krammer C I, Neto M F, Brielmann R M, Pedersen J S, Morimoto R I (2015) Investigating the spreading and toxicity of Prion-like proteins using the metazoan model organism C. elegans. J Vis Exp (95):52321. https://doi.org/10.3791/52321
Peng J -Y, Lin C -C, Chen Y -J, Kao L -S, Liu Y -C, Chou C -C, Huang Y -H, Chang F -R, Wu Y -C, Tsai Y -S, Hsu C -N (2011) Automatic morphological subtyping reveals new roles of Caspases in mitochondrial dynamics. PLOS Comput Biol 7(10):e1002212. https://doi.org/10.1371/journal.pcbi.1002212
Peng J-Y, Hsu C-N, Lin C-C (2010) Adaptive image enhancement for fluorescence microscopy. In: 2010 international conference on technologies and applications of artificial intelligence (TAAI). IEEE, pp 9–16. https://doi.org/10.1109/TAAI.2010.13
Otsu N (1979) A threshold selection method from gray-level histograms. IEEE T Syst Man Cyb 9(1):62–66. https://doi.org/10.1109/TSMC.1979.4310076
Tineveza J -Y, Perrya N, Schindelinb J, Hoopesc G M, Reynoldsc G D, Laplantined E, Bednarekc S Y, Shortea S L, Eliceiribe K W (2017) Trackmate: An open and extensible platform for single-particle tracking. Methods 115:80–90. https://doi.org/10.1016/j.ymeth.2016.09.016
Jaqaman K, Loerke D, Mettlen M, Kuwata H, Grinstein S, Schmid S L, Danuser G (2008) Robust single particle tracking in live cell time-lapse sequences. Nat Methods 5(8):695–702. https://doi.org/10.1038/nmeth.1237
Lantuejoul C, Beucher S (1981) On the use of the geodesic metric in image analysis. J Microsc 121(1):39–49. https://doi.org/10.1111/j.1365-2818.1981.tb01197.x
Ku T -C, Kao L -S, Lin C -C, Tsai Y -S (2009) Morphological filter improve the efficiency of automated tracking of secretory vesicles with various dynamic properties. Microsc Res Tech 72 (9):639–649. https://doi.org/10.1002/jemt.20711
Godinez W J, Lampe M, Wörz S, Müller B, Eils R, Rohr K (2009) Deterministic and probabilistic approaches for tracking virus particles in time-lapse fluorescence microscopy image sequences. Med Image Anal 13(2):325–42. https://doi.org/10.1016/j.media.2008.12.004
Tseng Y, Kole T P, Wirtz D (2002) Micromechanical mapping of live cells by multiple-particle-tracking microrheology. Biophys J 83(6):3162–3176. https://doi.org/10.1016/S0006-3495(02)75319-8
Apgar J, Tseng Y, Fedorov E, Herwig M B, Almo S C, Wirtz D (2000) Multiple-particle tracking measurements of heterogeneities in solutions of actin filaments and actin bundles. Biophys J 79(2):1095–1106. https://doi.org/10.1016/S0006-3495(00)76363-6
Jaeger S, Song Q, Chen S -S (2009) DYNAMIK: A Software environment for cell DYNAmics, motility, and information tracking, with an application to Ras pathways. Bioinformatics 25(18):2383–8. https://doi.org/10.1093/bioinformatics/btp405
Wang R, Perez-Riverol Y, Hermjakob H, Vizcaíno J A (2015) Open source libraries and frameworks for biological data visualisation: a guide for developers. Proteomics 15(8):1356–1374. https://doi.org/10.1002/pmic.201400377
Talevich E, Invergo B M, Cock P J A, Chapman B A (2012) Bio.phylo: a unified toolkit for processing, analyzing and visualizing phylogenetic trees in Biopython. BMC Bioinforma 13:209. https://doi.org/10.1186/1471-2105-13-209
Koskinen V R, Emery P A, Creasy D M, Cottrell J S (2011) Hierarchical clustering of shotgun proteomics data. Mol Cell Proteomics 003822(6):M110. https://doi.org/10.1074/mcp.M110.003822
Shneiderman B, Wattenberg M (2001) Ordered treemap layouts. In: IEEE Symposium on Information Visualization. INFOVIS https://doi.org/10.1109/INFVIS.2001.963283
Wang S, Xiao W, Shan S, Jiang C, Chen M, Zhang Y, Lü S, Chen J, Zhang C, Chen Q, Long M (2012) Multi-patterned dynamics of mitochondrial fission and fusion in a living cell. PLoS One 7(5):e19879. https://doi.org/10.1371/journal.pone.0019879
Dan H -W, Hung W -C, Tsai Y -S, Lin C -C (2014) 3D Image processing for mitochondria morphology variation analysis. In: Proceedings of the IEEE international symposium on bioelectronics and bioinformatics (ISBB). https://doi.org/10.1109/ISBB.2014.6820953
Dan (2020) Calculate blur kernel from original and blurry images https://www.mathworks.com/matlabcentral/fileexchange/54944-calculate-blur-kernel-from-original-and-blurry-images, MATLAB Central File Exchange: Retrieved April 15, 2020
Zhang Y (2020) 2D/3D image segmentation toolbox https://www.mathworks.com/matlabcentral/fileexchange/24998-2d-3d-image-segmentation-toolbox, MATLAB Central File Exchange. Retrieved April 15
Chan T F, Vese L A (2001) Active contours without edges. IEEE Trans Image Process 10 (2):266–277. https://doi.org/10.1109/83.902291
Achanta R, Hemami S, Estrada F, Susstrunk S (2009) Frequency-tuned salient region detection. In: Proceedings of the IEEE conference on computer vision and pattern recognition (CVPR). https://doi.org/10.1109/CVPR.2009.5206596
Fenga Y, Guoa H, Zhanga H, Lia C, Suna L, Muticc S, Jid S, Huc Y (2016) A modified fuzzy C-means method for segmenting MR images using non-local information. Technol Health Care 24:S785–S793. https://doi.org/10.1002/mp.12350
Feng Y, Dong F, Xia X, Hu C -H, Fan Q, Hu Y, Gao M, Mutic S (2017) An adaptive Fuzzy C-means method utilizing neighboring information for breast tumor segmentation in ultrasound images. Med Phys 44(7):3752–3760. https://doi.org/10.3233/THC-161208
Yang J -S, Valente C, Polishchuk R S, Turacchio G, Layre E, Moody D B, Leslie C C, Gelb M H, Brown W J, Corda D, Luini A, Hsu V W (2011) COPI Acts in both vesicular and tubular transport. Nat Cell Biol 13(8):996–1003. https://doi.org/10.1038/ncb2273
Funding
This research is partially supported by the Ministry of Science and Technology, Taiwan R.O.C. under Grant No. MOST 107-2221-E-033-023-MY2. LFH was supported by a postgraduate fellowship from the Irish Research Council (IRC).
Author information
Authors and Affiliations
Contributions
JCS and LFH collected the image data. Conceptualization and study design was performed by JY, YST, and CCL; methodology by JY and YST; implementation by JY; and validation by YST, CCL, and JCS. All authors interpreted the results data. JY, CCL, and JCS wrote the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Yaothak, J., Simpson, J.C., Heffernan, L.F. et al. 2D-GolgiTrack—a semi-automated tracking system to quantify morphological changes and dynamics of the Golgi apparatus and Golgi-derived membrane tubules. Med Biol Eng Comput 60, 151–169 (2022). https://doi.org/10.1007/s11517-021-02460-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11517-021-02460-5