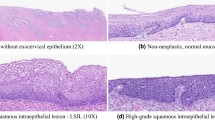
Abstract
The cervical cancer developing from the precancerous lesions caused by the human papillomavirus (HPV) has been one of the preventable cancers with the help of periodic screening. Cervical intraepithelial neoplasia (CIN) and squamous intraepithelial lesion (SIL) are two types of grading conventions widely accepted by pathologists. On the other hand, inter-observer variability is an important issue for final diagnosis. In this paper, a whole-slide image grading benchmark for cervical cancer precursor lesions is created and the “Uterine Cervical Cancer Database” introduced in this article is the first publicly available cervical tissue microscopy image dataset. In addition, a morphological feature representing the angle between the basal membrane (BM) and the major axis of each nucleus in the tissue is proposed. The presence of papillae of the cervical epithelium and overlapping cell problems are also discussed. Besides that, the inter-observer variability is also evaluated by thorough comparisons among decisions of pathologists, as well as the final diagnosis.













Similar content being viewed by others
References
Achanta R, Shaji A, Smith K, Lucchi A, Fua P, Süsstrunk S (2012) Slic superpixels compared to state-of-the-art superpixel methods. IEEE Trans Pattern Anal Mach Intell 34(11):2274–2282
Al-Janabi S, Huisman A, VDP J (2012) Digital pathology: current status and future perspectives. Histopathology 61(1):1–9
Arteta C, Lempitsky V, Noble JA, Zisserman A (2012) Learning to detect cells using non-overlapping extremal regions. In: International conference on medical image computing and computer-assisted intervention, Springer, pp 348–356
Cox JT, Wilkinson EJ, O’connor DM (2013) Historical perspective: terminology for lower anogenital tract pathology. AJSP: Rev Reports 18(4):158–167
Darragh TM, Colgan TJ, Cox JT, Heller DS, Henry MR, Luff RD, McCalmont T, Nayar R, Palefsky JM, Stoler MH et al (2013) The lower anogenital squamous terminology standardization project for hpv-associated lesions: background and consensus recommendations from the college of american pathologists and the american society for colposcopy and cervical pathology. Int J Gynecol Pathol 32(1):76–115
Darragh TM, et al. (2012) The lower anogenital squamous terminology standardization project for hpv-associated lesions: background and consensus recommendations from the college of american pathologists and the american society for colposcopy and cervical pathology. Arch Path Lab Med 136(10):1266–1297
De S, Stanley RJ, Lu C, Long R, Antani S, Thoma G, Zuna R (2013) A fusion-based approach for uterine cervical cancer histology image classification. Comput Med Imaging Graph 37(7):475–487
Doorbar J (2007) Papillomavirus life cycle organization and biomarker selection. Dis Markers 23 (4):297–313
Galgano MT, Castle PE, Atkins KA, Brix WK, Nassau SR, Stoler MH (2010) Using biomarkers as objective standards in the diagnosis of cervical biopsies. Am J Surg Path 34(8):1077
Guo M, Baruch A, Silva E, Jan Y, Lin E, Sneige N, Deavers MT (2011) Efficacy of p16 and proexc immunostaining in the detection of high-grade cervical intraepithelial neoplasia and cervical carcinoma. Am J Clin Pathol 135(2):212–220
Guo P, Banerjee K, Stanley RJ, Long R, Antani S, Thoma G, Zuna R, Frazier SR, Moss RH, Stoecker WV (2016) Nuclei-based features for uterine cervical cancer histology image analysis with fusion-based classification. IEEE J Biomed Health Inform 20(6):1595–1607
He L, Long LR, Antani S, Thoma GR (2012) Histology image analysis for carcinoma detection and grading. Comput Methods Programs Biomed 107(3):538–556
Hong X, Chen S, Harris CJ (2007) A kernel-based two-class classifier for imbalanced data sets. IEEE Trans Neur Netw 18(1):28–41
Jordan J, Arbyn M, Martin-Hirsch P, Schenck U, Baldauf JJ, Da Silva D, Anttila A, Nieminen P, Prendiville W (2008) European guidelines for quality assurance in cervical cancer screening: recommendations for clinical management of abnormal cervical cytology, part 1. Cytopathology 19(6):342–354
Keenan S, Diamond J, McCluggage G, Bharucha H, Thompson D, Bartels PH, Hamilton PW (2000) An automated machine vision system for the histological grading of cervical intraepithelial neoplasia (cin). J Pathol 192(3):351–362
Kurman RJ (2013) Blaustein’s pathology of the female genital tract. Springer Science & Business Media, Berlin
Lim S, Lee M, Cho I, Hong R, Lim S (2016) Efficacy of p16 and ki-67 immunostaining in the detection of squamous intraepithelial lesions in a high-risk hpv group. Oncol Lett 11(2):1447–1452
Liu W, Chawla S (2011) Class confidence weighted knn algorithms for imbalanced data sets. In: Pacific-asia conference on knowledge discovery and data mining, Springer, pp 345–356
Madabhushi A, Lee G (2016) Image analysis and machine learning in digital pathology: Challenges and opportunities. Med Image Anal 33:170–175
Mazurowski MA, Habas PA, Zurada JM, Lo JY, Baker JA, Tourassi GD (2008) Training neural network classifiers for medical decision making: The effects of imbalanced datasets on classification performance. Neural Netw 21(2-3):427–436
McCluggage W, Bharucha H, Caughley L, Date A, Hamilton P, Thornton C, Walsh M (1996) Interobserver variation in the reporting of cervical colposcopic biopsy specimens: Comparison of grading systems. J Clin Path 49(10):833–835
McCluggage W, Walsh M, Thornton C, Hamilton P, Caughley L, Bharucha H, et al. (1998) Inter-and intra-observer variation in the histopathological reporting of cervical squamous in traepithelial lesion susing a modified bethesda grading system. Br J Obstet Gynaecol 105(2):206–210
de Melo F, Lancellotti C, da Silva M (2016) 16 and ki-67 and their usefulness in the diagnosis of cervicalexpression of the immunohistochemical markers p intraepithelial neoplasms. Rev Bras Ginicol Obstet 38(2):82–87
Miranda G H B, Soares E G, Barrera J, Felipe JC (2012) Method to support diagnosis of cervical intraepithelial neoplasia (cin) based on structural analysis of histological images. In: 2012 25th IEEE International symposium on computer-based medical systems (CBMS) IEEE, pp 1–6
Mitra A, MacIntyre D, Lee Y, Smith A, Marchesi JR, Lehne B, Bhatia R, Lyons D, Paraskevaidis E, Li J et al (2015) Cervical intraepithelial neoplasia disease progression is associated with increased vaginal microbiome diversity. Sci Rep 5:16,865
Naghdy G, Ros MB, Todd C et al (2012) Computer aided decision support system for cervical cancer classification. In: Applications of Digital Image Processing XXXV, SPIE, vol 8499, p 849919
Najar F, Bourouis S, Bouguila N, Belghith S (2017) A comparison between different gaussian-based mixture models. In: 2017 IEEE/ACS 14th International conference on computer systems and applications, AICCSA’17, IEEE, pp 704–708
Nayar R, Wilbur DC (2015) The Bethesda system for reporting cervical cytology: definitions, criteria, and explanatory notes. Springer, Berlin
Ozaki S, Zen Y, Inoue M (2011) Biomarker expression in cervical intraepithelial neoplasia: potential progression predictive factors for low-grade lesions. Hum Pathol 42(7):1007–1012
Park KJ, Soslow RA (2009) Current concepts in cervical pathology. Arch Pathol Lab Med 133(5):729–738
Qi X, Xing F, Foran D J, Yang L (2011) Robust segmentation of overlapping cells in histopathology specimens using parallel seed detection and repulsive level set. IEEE Trans Biomed Eng 59(3):754–765
Stoler MH (2000a) Advances in cervical screening technology. Mod Pathol 13(3):275–284
Stoler MH (2000b) Human papillomaviruses and cervical neoplasia: a model for carcinogenesis. Int J Gynecol Pathol 19(1):16–28
Stoler MH, Schiffman M, et al. (2001) Interobserver reproducibility of cervical cytologic and histologic interpretations: realistic estimates from the ascus-lsil triage study. Jama 285(11):1500–1505
Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A (2012) Global cancer statistics. CA Cancer J Clin 65(2):87–108
Van Zummeren M, Kremer WW, Leeman A, Bleeker MC, Jenkins D, van de Sandt M, Doorbar J, Heideman DA, Steenbergen RD, Snijders PJ et al (2018) Hpv e4 expression and dna hypermethylation of cadm1, mal, and mir124-2 genes in cervical cancer and precursor lesions. Mod Pathol 1
Crookes D, Wang Y, Eldin OS, Hamilton P, Wang S, Diamond J (2009) Assisted diagnosis of cervical intraepithelial neoplasia (cin). IEEE J Sel Top Signal Process 3(1):112–121
Zhuang L, Dai H (2006) Parameter optimization of kernel-based one-class classifier on imbalance learning. J Comput 1(7):32–40
Zuo W, Lu W, Wang K, Zhang H (2008) Diagnosis of cardiac arrhythmia using kernel difference weighted knn classifier. In: 2008 Computers in cardiology, IEEE, pp 253–256
Zur HH (2009) Papillomaviruses in the causation of human cancers—a brief historical account. Virol J 384(2):260–265
Acknowledgements
The authors also would like to thank Argenit Company and Istanbul Medipol University Hospital for providing and annotating the whole-slide histopathological images of cervical cancer precursor lesions image dataset. The authors would like to thank the reviewers for all useful and instructive comments on our manuscript.
Funding
This work is in part funded by ITU BAP MAB-2020-42314 project and also supported by the Scientific Research Projects Coordination Department, Yildiz Technical University, under Project 2014-04-01-KAP01.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Albayrak, A., Akhan, A.U., Calik, N. et al. A whole-slide image grading benchmark and tissue classification for cervical cancer precursor lesions with inter-observer variability. Med Biol Eng Comput 59, 1545–1561 (2021). https://doi.org/10.1007/s11517-021-02388-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11517-021-02388-w