Abstract
In this work, we propose a heterogeneous committee (ensemble) of diverse members (classification approaches) to solve the problem of human epithelial (HEp-2) cell image classification using indirect Immunofluorescence (IIF) imaging. We hypothesize that an ensemble involving different feature representations can enable higher performance if individual members in the ensemble are sufficiently varied. These members are of two types: (1) CNN-based members, (2) traditional members. For the CNN members, we have employed the well-established ResNet, DenseNet, and Inception models, which have distinctive salient aspects. For the traditional members, we incorporate class-specific features which are characterized depending on visual morphological attributes, and some standard texture features. To select the members which are discriminating and not redundant, we use an information theoretic measure which considers the trade-off between individual accuracies and diversity among the members. For all selected members, a compelling fusion required to combine their outputs to reach a final decision. Thus, we also investigate various fusion methods that combine the opinion of the committee at different levels: maximum voting, product, decision template, Bayes, Dempster-Shafer, etc. The proposed method is evaluated using ICPR-2014 data which consists of more images than some previous datasets ICPR-2012 and demonstrate state-of-the-art performance. To check the effectiveness of the proposed methodology for other related datasets, we test our methodology with newly compiled large-scale HEp-2 dataset with 63K cell images and demonstrate comparable performance even with less number of training samples. The proposed method produces 99.80% and 86.03% accuracy respectively when tested on ICPR-2014 and a new large-scale data containing 63K samples.

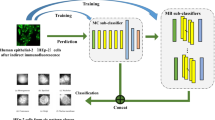
Overview of the proposed methodology






Similar content being viewed by others
References
Wiik AS, Høier-madsen M, Forslid J, Charles P, Meyrowitsch J (2010) Antinuclear antibodies: a contemporary nomenclature using HEp-2 cells. J Autoimmun 35(3):276–290
Meroni PL, Schur PH (2010) ANA screening: an old test with new recommendations. Ann Rheum Diseases 69(8):1420–1422
Kumar Y, Bhatia A, Minz RW (2009) Antinuclear antibodies and their detection methods in diagnosis of connective tissue diseases: a journey revisited. Diag Path 4(1):1–10
Hiemann R, Büttner T, Krieger T, Roggenbuck D, Sack U, Conrad K (2009) Challenges of automated screening and differentiation of non-organ specific autoantibodies on HEp-2 cells. Autoimmun Rev 9(1):17–22
Sack U, Knoechner S, Warschkau H, Pigla U, Emmrich F, Kamprad M (2003) Computer-assisted classification of HEp-2 immunofluorescence patterns in autoimmune diagnostics. Autoimmun Rev 2 (5):298–304
Hiemann R, Hilger N, Sack U, Weigert M (2006) Objective quality evaluation of fluorescence images to optimize automatic image acquisition. Cytometry Part A 69(3):182–184
Pham B, Albarede S, Guyard A, Burg E, Maisonneuve P (2005) Impact of external quality assessment on antinuclear antibody detection performance. Lupus 14(2):113–119
Bell DA, Guan JwW, Bi Y (2005) On combining classifier mass functions for text categorization. IEEE Trans Knowl Data Eng 17(10):1307–1319
Wolpert DH (1992) Stacked generalization. Neural Netw 5(2):241–259
Pajares G, Guijarro M, Ribeiro A (2010) A hopfield neural network for combining classifiers applied to textured images. Neural Netw 23(1):144–153
Su Y, Shan S, Chen X, Gao W (2009) Hierarchical ensemble of global and local classifiers for face recognition. IEEE Trans Image Process 18(8):1885–1896
Krizhevsky A, Sutskever I, Hinton GE (2012) Imagenet classification with deep convolutional neural networks. In: Advances in neural information processing systems, pp 1097–1105
Ponti MP Jr (2011) Combining classifiers: from the creation of ensembles to the decision fusion. In: 2011 24th SIBGRAPI conference on graphics, patterns and images tutorials (SIBGRAPI-T). IEEE, pp 1–10
Wolpert DH (2002) The supervised learning no-free-lunch theorems. In: Soft computing and industry. Springer, pp 25–42
Breiman L (2001) Random forests. Mach Learn 45(1):5–32
Sharif Razavian A, Azizpour H, Sullivan J, Carlsson S (2014) CNN features off-the-shelf: an astounding baseline for recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition workshops, pp 806–813
Shen H, Manivannan S, Annunziata R, Wang R, Zhang J (2016) Combination of CNN and hand-crafted feature for Ischemic stroke lesion segmentation. Ischemic Stroke Lesion Segment: 1
Li W, Manivannan S, Akbar S, Zhang J, Trucco E, McKenna SJ (2016) Gland segmentation in colon histology images using hand-crafted features and convolutional neural networks. In: 2016 IEEE 13th international symposium on biomedical imaging (ISBI). IEEE, pp 1405–1408
Meynet J, Thiran JP (2010) Information theoretic combination of pattern classifiers. Patt Recogn 43(10):3412–3421
Kuncheva LI, Whitaker CJ (2003) Measures of diversity in classifier ensembles and their relationship with the ensemble accuracy. Mach Learn 51(2):181–207
Brown G, Wyatt J, Harris R, Yao X (2005) Diversity creation methods: a survey and categorisation. Inf Fus 6(1):5–20
Qi X, Zhao G, Chen J, Pietikäinen M (2016) Exploring illumination robust descriptors for human epithelial type 2 cell classification. Patt Recogn 60:420–429
Percannella G, Foggia P, Soda P (2012) Contest on HEp-2 cells classification. In: IEEE international conference on pattern recognition (ICPR)
Wiliem A, Wong Y, Sanderson C, Hobson P, Chen S, Lovell BC (2013) Classification of human epithelial type 2 cell indirect immunofluoresence images via codebook based descriptors. In: 2013 IEEE workshop on applications of computer vision (WACV). IEEE, pp 95–102
Hobson P, Lovell BC, Percannella G, Vento M, Wiliem A (2015) Benchmarking human epithelial type 2 interphase cells classification methods on a very large dataset. Artif Intell Med 65(3):239–250
Lovell B, Percannella G, Vento M, Wiliem A (2014) 1st workshop on pattern recognition techniques for indirect immunofluorescence images. In: IEEE international conference on pattern recognition (ICPR)
Foggia P, Percannella G, Soda P, Vento M (2010) Early experiences in mitotic cells recognition on HEp-2 slides. In: 2010 IEEE 23rd international symposium on computer-based medical systems (CBMS). IEEE, pp 38–43
Iannello G, Percannella G, Soda P, Vento M (2014) Mitotic cells recognition in HEp-2 images. Patt Recogn Lett 45:136–144
Hobson P, Lovell BC, Percannella G, Saggese A, Vento M, Wiliem A (2016) HEP-2 staining pattern recognition at cell and specimen levels: datasets, algorithms and results. Patt Recogn Lett 82:12–22
Hobson P, Lovell BC, Percannella G, Saggese A, Vento M, Wiliem A (2016) Computer aided diagnosis for anti-nuclear antibodies HEp-2 images: progress and challenges. Patt Recogn Lett 82:3–11
Cascio D, Taormina V, Cipolla M, Bruno S, Fauci F, Raso G (2016) A multi-process system for HEp-2 cells classification based on SVM. Patt Recogn Lett 82:56–63
Ensafi S, Lu S, Kassim AA, Tan CL (2016) Accurate HEp-2 cell classification based on sparse coding of superpixels. Patt Recogn Lett 82:64–71
Han XH, Chen YW, Xu G (2016) Integration of spatial and orientation contexts in local ternary patterns for HEp-2 cell classification. Patt Recogn Lett 82:23–27
Gragnaniello D, Sansone C, Verdoliva L (2016) Cell image classification by a scale and rotation invariant dense local descriptor. Patt Recogn Lett 82:72–78
Nanni L, Lumini A, dos Santos FLC, Paci M, Hyttinen J (2016) Ensembles of dense and dense sampling descriptors for the HEp-2 cells classification problem. Patt Recogn Lett 82:28–35
Chandran V, Banks J, Boles W, Chen B, Tomeo-Reyes I (2013) Cell image classification using histograms, higher order statistics and adaboost. In: IEEE International Conference on Image Processing (ICIP) - Cells Classification by Fluorescent Image Analysis Competition: 11
Nanni L, Paci M, dos Santos FLC, Hyttinen J (2014) Morphological and texture features for HEp-2 cells classification. In: 1st workshop on pattern recognition techniques for indirect immunofluorescence images (I3A). IEEE, pp 45–48
Theodorakopoulos I, Kastaniotis D, Economou G, Fotopoulos S (2014) HEp-2 cells classification using morphological features and a bundle of local gradient descriptors. In: 1st workshop on pattern recognition techniques for indirect immunofluorescence images (I3A). IEEE, pp 33–36
Theodorakopoulos I, Kastaniotis D, Economou G, Fotopoulos S (2012) HEp-2 cells classification via fusion of morphological and textural features. In: 12th international conference on bioinformatics & bioengineering (BIBE). IEEE, pp 689–694
Donato C, Vincenzo T, Marco C, Francesco F, Maria VS, Giuseppe R (2014) HEp-2 cell classification with heterogeneous classes-processes based on k-nearest neighbours. In: 1st workshop on pattern recognition techniques for indirect immunofluorescence images (I3A). IEEE, pp 10–15
Ensafi S, Lu S, Kassim AA, Tan CL (2014) A bag of words based approach for classification of HEp-2 cell images. In: 1st workshop on pattern recognition techniques for indirect immunofluorescence images (I3A). IEEE, pp 29–32
Schapire RE (1990) The strength of weak learnability. Mach Learn 5(2):197–227
Freund Y, Schapire RE et al (1996) Experiments with a new boosting algorithm. In: icml, vol 96, pp 148–156
Manivannan S, Li W, Akbar S, Wang R, Zhang J, McKenna SJ (2014) HEp-2 cell classification using multi-resolution local patterns and ensemble SVMs. In: 1st workshop on pattern recognition techniques for indirect immunofluorescence images (I3A). IEEE, pp 37–40
Manivannan S, Li W, Akbar S, Wang R, Zhang J, McKenna SJ (2016) An automated pattern recognition system for classifying indirect immunofluorescence images of HEp-2 cells and specimens. Patt Recogn 51:12–26
Gao Z, Zhang J, Zhou L, Wang L (2014) Hep-2 cell image classification with convolutional neural networks. In: 2014 1st workshop on pattern recognition techniques for indirect immunofluorescence images (I3A). IEEE, pp 24–28
Gao Z, Wang L, Zhou L, Zhang J (2017) HEP-2 cell image classification with deep convolutional neural networks. IEEE J Biomed Health Inf 21(2):416–428
Rodrigues LF, Naldi MC, Mari JF (2017) Exploiting Convolutional Neural Networks and preprocessing techniques for HEp-2 cell classification in immunofluorescence images. In: 2017 30th SIBGRAPI conference on graphics, patterns and images (SIBGRAPI). IEEE, pp 170–177
Jia X, Shen L, Zhou X, Yu S (2016) Deep convolutional neural network based HEp-2 cell classification. In: IEEE, pp 77–80
Li Y, Shen L (2018) HEP-net: a smaller and better deep-learning network for HEp-2 cell classification. Computer Methods in Biomechanics and Biomedical Engineering: Imaging & Visualization 7(3):1–7
Phan HTH, Kumar A, Kim J, Feng D (2016) Transfer learning of a convolutional neural network for HEp-2 cell image classification. In: 2016 IEEE 13th international symposium on biomedical imaging (ISBI). IEEE, pp 1208–1211
Bayramoglu N, Kannala J, Heikkilä J (2015) Human epithelial type 2 cell classification with convolutional neural networks. In: 2015 IEEE 15th international conference on bioinformatics and bioengineering (BIBE). IEEE, pp 1–6
Liu J, Xu B, Shen L, Garibaldi J, Qiu G (2017) HEp-2 cell classification based on a deep autoencoding-classification convolutional neural network. In: 2017 IEEE 14th international symposium on biomedical imaging (ISBI 2017). IEEE, pp 1019–1023
Wen G, Hou Z, Li H, Li D, Jiang L, Xun E (2017) Ensemble of deep neural networks with probability-based fusion for facial expression recognition. Cogn Comput 9(5):597–610
Kim BK, Roh J, Dong SY, Lee SY (2016) Hierarchical committee of deep convolutional neural networks for robust facial expression recognition. J Multimodal User Interf 10(2):173–189
Gupta V, Gupta K, Bhavsar A, Sao AK (2016) Hierarchical classification of HEp-2 cell images using class-specific features. In: 2016 6th European workshop on visual information processing (EUVIP), vol 7, no. 3. IEEE, pp 1–6
Gupta K, Gupta V, Sao AK, Bhavsar A, Dileep AD (2014) Class-specific hierarchical classification of HEp-2 cell images: the case of two classes. In: 2014 1st workshop on pattern recognition techniques for indirect immunofluorescence images. IEEE , pp 6–9
Szegedy C, Liu W, Jia Y, Sermanet P, Reed S, Anguelov D, et al. (2015) Going deeper with convolutions. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 1–9
Huang G, Liu Z, Van Der Maaten L, Weinberger KQ (2017) Densely connected convolutional networks. In: CVPR, vol 1, p 3
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 770–778
Strandmark P, Ulén J, Kahl F (2012) HEp-2 staining pattern classification. In: 21st international conference on pattern recognition (ICPR). IEEE, pp 33–36
Chang CC, Lin CJ (2011) LIBSVM: A library for support vector machines. ACM Trans Intell Sys Technol (TIST) 2(3):27
Chen T, Guestrin C (2016) XGBoost: reliable large-scale tree boosting system. arXiv. 2016a ISSN. pp 0146–4833
Kuncheva LI, Whitaker CJ (2002) Using diversity with three variants of boosting: aggressive, conservative, and inverse. In: International workshop on multiple classifier systems. Springer, pp 81–90
Xu L, Krzyzak A, Suen CY (1992) Methods of combining multiple classifiers and their applications to handwriting recognition. IEEE Trans Sys Man Cybern 22(3):418–435
Kuncheva LI (2004) Combining pattern classifiers: methods and algorithms. Wiley, New York
Singh D, Singh B (2019) Investigating the impact of data normalization on classification performance. Appl Soft Comput 97 Part B:105524
Erhan D, Manzagol PA, Bengio Y, Bengio S, Vincent P (2009) The difficulty of training deep architectures and the effect of unsupervised pre-training. In: Artificial Intelligence and Statistics, pp 153–160
Margeta J, Criminisi A, Cabrera Lozoya R, Lee DC, Ayache N (2017) Fine-tuned convolutional neural nets for cardiac MRI acquisition plane recognition. Comput Methods Biomech Biomed Eng Imag Visual 5(5):339–349
Kumar A, Sridar P, Quinton A, Kumar RK, Feng D, Nanan R et al (2016) Plane identification in fetal ultrasound images using saliency maps and convolutional neural networks. In: 2016 IEEE 13th international symposium on biomedical imaging (ISBI). IEEE, pp 791–794
Bayramoglu N, Heikkilä J (2016) Transfer learning for cell nuclei classification in histopathology images. In: European conference on computer vision. Springer, pp 532–539
Penatti OA, Nogueira K, dos Santos JA (2015) Do deep features generalize from everyday objects to remote sensing and aerial scenes domains?. In: Proceedings of the IEEE conference on computer vision and pattern recognition workshops, pp 44–51
Azizpour H, Sharif Razavian A, Sullivan J, Maki A, Carlsson S (2015) From generic to specific deep representations for visual recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition workshops, pp 36–45
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Author contributions
The first author was primarily responsible for the work reported in this paper. The second author played an advisory role involving various discussions. The second author is the Ph.D. advisor of the first author.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Gupta, V., Bhavsar, A. Heterogeneous ensemble with information theoretic diversity measure for human epithelial cell image classification. Med Biol Eng Comput 59, 1035–1054 (2021). https://doi.org/10.1007/s11517-021-02336-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11517-021-02336-8