Abstract
Pain is a significant public health problem, affecting millions of people in the USA. Evidence has highlighted that patients with chronic pain often suffer from deficits in pain care quality (PCQ) including pain assessment, treatment, and reassessment. Currently, there is no intelligent and reliable approach to identify PCQ indicators inelectronic health records (EHR). Hereby, we used unstructured text narratives in the EHR to derive pain assessment in clinical notes for patients with chronic pain. Our dataset includes patients with documented pain intensity rating ratings > = 4 and initial musculoskeletal diagnoses (MSD) captured by (ICD-9-CM codes) in fiscal year 2011 and a minimal 1 year of follow-up (follow-up period is 3-yr maximum); with complete data on key demographic variables. A total of 92 patients with 1058 notes was used. First, we manually annotated qualifiers and descriptors of pain assessment using the annotation schema that we previously developed. Second, we developed a reliable classifier for indicators of pain assessment in clinical note. Based on our annotation schema, we found variations in documenting the subclasses of pain assessment. In positive notes, providers mostly documented assessment of pain site (67%) and intensity of pain (57%), followed by persistence (32%). In only 27% of positive notes, did providers document a presumed etiology for the pain complaint or diagnosis. Documentation of patients’ reports of factors that aggravate pain was only present in 11% of positive notes. Random forest classifier achieved the best performance labeling clinical notes with pain assessment information, compared to other classifiers; 94, 95, 94, and 94% was observed in terms of accuracy, PPV, F1-score, and AUC, respectively. Despite the wide spectrum of research that utilizes machine learning in many clinical applications, none explored using these methods for pain assessment research. In addition, previous studies using large datasets to detect and analyze characteristics of patients with various types of pain have relied exclusively on billing and coded data as the main source of information. This study, in contrast, harnessed unstructured narrative text data from the EHR to detect pain assessment clinical notes. We developed a Random forest classifier to identify clinical notes with pain assessment information. Compared to other classifiers, ours achieved the best results in most of the reported metrics.

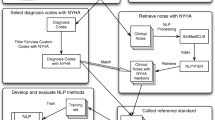
Framework for detecting pain assessment in clinical notes.


Similar content being viewed by others
References
Simon LS (2012) Relieving pain in America: a blueprint for transforming prevention, care, education, and research. J Pain Palliative Care Pharmacother 26(2):197–198
Dorflinger LM, Gilliam WP, Lee AW, Kerns RD (2014) Development and application of an electronic health record information extraction tool to assess quality of pain management in primary care. Transl Behav Med 4(2):184–189
Hooten W, Timming R, Belgrade M. Assessment and management of chronic pain. Bloomington, MN: Institute for Clinical Systems Improvement; 2013. h ttps
Tian TY, Zlateva I, Anderson DR (2013) Using electronic health records data to identify patients with chronic pain in a primary care setting. J Am Med Inform Assoc 20(e2):e275–e280
Sinnott PL, Siroka AM, Shane AC, Trafton JA, Wagner TH (2012) Identifying neck and back pain in administrative data: defining the right cohort. Spine 37(10):860–874
Plaisance L (2000) Pain—Clinical Manual. Home Healthcare Now 18(8):556
Krebs EE, Carey TS, Weinberger M (2007) Accuracy of the pain numeric rating scale as a screening test in primary care. J Gen Intern Med 22(10):1453–1458
Goetzke G, Johns T, Reid M, Borg J (2001) Carlson a. Chronic pain patient identification system, Google Patents
Maeng DD, Stewart WF, Yan X et al (2015) Use of electronic health records for early detection of high-cost, low back pain patients. Pain Res Manag 20(5):234–240
Jordan KP, Timmis A, Croft P et al (2017) Prognosis of undiagnosed chest pain: linked electronic health record cohort study. BMJ 357:j1194
Bui DDA, Zeng-Treitler Q (2014) Learning regular expressions for clinical text classification. J Am Med Inform Assoc 21(5):850–857
Sellinger JJ, Wallio SC, Clark EA, Kerns RD, Ebert M, Kerns R (2010) Comprehensive pain assessment: the integration of biopsychosocial principles. Cambridge University Press New York
Anderson D, Zlateva I, Lee A, Tian T, Khatri K, Ruser CB (2016) Stepped care model for pain management and quality of pain care in long-term opioid therapy. J Rehabil Res Dev 53(1):137
Haskell SG, Brandt CA, Krebs EE, Skanderson M, Kerns RD, Goulet JL (2009) Pain among veterans of operations enduring freedom and Iraqi freedom: do women and men differ? Pain Med 10(7):1167–1173
Affairs DoV. Chapter 264: pact primary care clinic (PPCC). 2015
Weed LL (1964) Medical records, patient care, and medical education. Irish J Med Sci (1926–1967) 39(6):271–282
Cameron S, Turtle-Song I (2002) Learning to write case notes using the SOAP format. J Couns Dev 80(3):286–292
South BR, Shen S, Leng J, Forbush TB, DuVall SL, Chapman WW (2012) A prototype tool set to support machine-assisted annotation. Paper presented at: Proceedings of the 2012 Workshop on Biomedical Natural Language Processing
Hripcsak G, Rothschild AS (2005) Agreement, the f-measure, and reliability in information retrieval. J Am Med Inform Assoc 12(3):296–298
Hripcsak G, Heitjan DF (2002) Measuring agreement in medical informatics reliability studies. J Biomed Inform 35(2):99–110
Ogren PV, Savova G, Buntrock JD, Chute CG (2006) Building and evaluating annotated corpora for medical NLP systems. Paper presented at: AMIA Annual Symposium Proceedings
Ogren PV, Savova GK, Chute CG. Constructing evaluation corpora for automated clinical named entity recognition. Paper presented at: Medinfo 2007: Proceedings of the 12th World Congress on Health (Medical) Informatics; Building Sustainable Health Systems2007
Meystre SM, Savova GK, Kipper-Schuler KC, Hurdle JF (2008) Extracting information from textual documents in the electronic health record: a review of recent research. Yearb Med Inform 35(128):44
SKI Learn. http://scikit-learn.org/stable/tutorial/index.html . 2016
Bird S. NLTK: the natural language toolkit. Paper presented at: Proceedings of the COLING/ACL on Interactive presentation sessions2006
Cunningham P, Delany SJ (2007) K-nearest neighbour classifiers. Multiple Classifier Syst 34:1–17
Peterson LE (2009) K-nearest neighbor. Scholarpedia 4(2):1883
Quinlan JR (1986) Induction of decision trees. Mach Learn 1(1):81–106
Safavian SR, Landgrebe D (1991) A survey of decision tree classifier methodology. IEEE Trans Syst Man Cybernetics 21(3):660–674
Xia F, Zhang W, Li F, Yang Y (2008) Ranking with decision tree. Knowl Inf Syst 17(3):381–395
Gunn SR (1998) Support vector machines for classification and regression. ISIS Tech Report 14:85–86
Chang C-C, Lin C-J (2011) LIBSVM: a library for support vector machines. ACM Trans Intell Syst Technol (TIST) 2(3):27
Svetnik V, Liaw A, Tong C, Culberson JC, Sheridan RP, Feuston BP (2003) Random forest: a classification and regression tool for compound classification and QSAR modeling. J Chem Inf Comput Sci 43(6):1947–1958
Liaw A, Wiener M (2002) Classification and regression by randomForest. R News 2(3):18–22
Kohavi R. A study of cross-validation and bootstrap for accuracy estimation and model selection. Paper presented at: Ijcai1995
Refaeilzadeh P, Tang L, Liu H. Cross-validation. Encyclopedia of database systems: Springer; 2009:532–538
Zhang P (1993) Model selection via multifold cross validation. Ann Stat:299–313
Dietterich TG (1998) Approximate statistical tests for comparing supervised classification learning algorithms. Neural Comput 10(7):1895–1923
Bouckaert RR. Choosing between two learning algorithms based on calibrated tests. Paper presented at: Proceedings of the 20th International Conference on Machine Learning (ICML-03)2003
Ross KA (2009) Cache-conscious query processing. Springer, Encyclopedia of Database Systems, pp 301–304
Fodeh SJ, Trentalange M, Allore HG, Gill TM, Brandt CA, Murphy TE (2015) Baseline cluster membership demonstrates positive associations with first occurrence of multiple gerontologic outcomes over 10 years. Exp Aging Res 41(2):177–192
Begg RK, Palaniswami M, Owen B (2005) Support vector machines for automated gait classification. IEEE Trans Biomed Eng 52(5):828–838
Widjaja E, Zheng W, Huang Z (2008) Classification of colonic tissues using near-infrared Raman spectroscopy and support vector machines. Int J Oncol 32(3):653–662
Orrù G, Pettersson-Yeo W, Marquand AF, Sartori G, Mechelli A (2012) Using support vector machine to identify imaging biomarkers of neurological and psychiatric disease: a critical review. Neurosci Biobehav Rev 36(4):1140–1152
El-Naqa I, Yang Y, Wernick MN, Galatsanos NP, Nishikawa RM (2002) A support vector machine approach for detection of microcalcifications. IEEE Trans Med Imaging 21(12):1552–1563
Lee Y, Lee C-K (2003) Classification of multiple cancer types by multicategory support vector machines using gene expression data. Bioinformatics 19(9):1132–1139
Pakhomov SV, Hanson PL, Bjornsen SS, Smith SA (2008) Automatic classification of foot examination findings using clinical notes and machine learning. J Am Med Inform Assoc 15(2):198–202
McCart JA, Berndt DJ, Finch D, Jarman J, Luther S. Using Statistical Text Mining to Identify Falls in VHA Ambulatory Care Data. Paper presented at: AMIA2012
Fodeh S, Benin A, Miller P, Lee K, Koss M, Brandt C. Laplacian SVM Based Feature Selection Improves Medical Event Reports Classification. Paper presented at: 2015 I.E. International Conference on Data Mining Workshop (ICDMW)2015
Cicero TJ, Wong G, Tian Y, Lynskey M, Todorov A, Isenberg K. Co-morbidity and utilization of medical services by pain patients receiving opioid medications: data from an insurance claims database. PAIN®. 2009;144(1):20–27
Breen AC, Carr E, Langworthy JE, Osmond C, Worswick L (2011) Back pain outcomes in primary care following a practice improvement intervention:-a prospective cohort study. BMC Musculoskelet Disord 12(1):1
Berger A, Sadosky A, Dukes E, Edelsberg J, Oster G (2012) Clinical characteristics and patterns of healthcare utilization in patients with painful neuropathic disorders in UK general practice: a retrospective cohort study. BMC Neurol 12(1):1
Sullivan MD, Edlund MJ, Fan M-Y, DeVries A, Braden JB, Martin BC (2010) Risks for possible and probable opioid misuse among recipients of chronic opioid therapy in commercial and medicaid insurance plans: the TROUP study. Pain 150(2):332–339
Goulet JL, Kerns RD, Bair M et al (2016) The musculoskeletal diagnosis cohort: examining pain and pain care among veterans. Pain 157(8):1696–1703
Moore BA, Anderson D, Dorflinger L, Zlateva I, Lee A, Gilliam W, Tian T, Khatri K, Ruser C, Kerns RD (2016) The stepped care model of pain management and quality of pain care in long-term opioid therapy. J Rehab Res Develop 53(1):137–146
Dworkin RH, Turk DC, Farrar JT, Haythornthwaite JA, Jensen MP, Katz NP, Kerns RD et al (2016) Core outcome measures for chronic pain clinical trials: IMMPACT recommendations. Pain 113:9–19
Funding
This study was funded by NIH National Center for Complementary and Alternative Medicine—grant number (1R01AT008448-01). It was also partially funded by the Veterans Affairs.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Fodeh, S.J., Finch, D., Bouayad, L. et al. Classifying clinical notes with pain assessment using machine learning. Med Biol Eng Comput 56, 1285–1292 (2018). https://doi.org/10.1007/s11517-017-1772-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11517-017-1772-1