Abstract
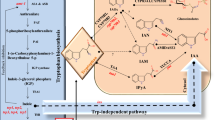
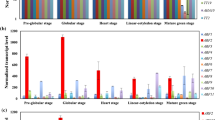
Both tryptophan (Trp) and auxin are essential for plant growth and Trp is a precursor for auxin biosynthesis. Concentrations of Trp and auxin need to be tightly controlled to ensure optimal growth and development. It has been very difficult to study the homeostasis of these two essential and inter-dependent compounds. Auxin is mainly synthesized from Trp via a two-step pathway using indole-3-pyruvate (IPA) as the intermediate. Here we used a bacterial Trp oxidase RebO, which does not exist in Arabidopsis and which converts Trp to the imine form of IPA, to modulate IPA levels in Arabidopsis. Our results demonstrate that Arabidopsis plants use two strategies to ensure that no excess IPA is made from Trp. IPA is made from Trp by the TAA family of aminotransferases, which we show catalyzes the reverse reaction when IPA level is high. Moreover, excess IPA is converted back to Trp by the VAS1 aminotransferase. We show that the VAS1-catalyzed reaction is very important for Trp homeostasis. This work not only elucidates the intricate biochemical mechanisms that control the homeostasis of Trp, IPA, and auxin, but also provides novel tools for further biochemical studies on Trp metabolism and auxin biosynthesis in plants.





Similar content being viewed by others
References
Zhao Y (2010) Auxin biosynthesis and its role in plant development. Annu Rev Plant Biol 61:49–64
Zhao Y (2014) Auxin biosynthesis. The Arabidopsis book 12:e0173
Cheng Y, Dai X, Zhao Y (2006) Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev 20:1790–1799
Cheng Y, Dai X, Zhao Y (2007) Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell 19:2430–2439
Pinon V, Prasad K, Grigg SP et al (2013) Local auxin biosynthesis regulation by PLETHORA transcription factors controls phyllotaxis in Arabidopsis. Proc Natl Acad Sci USA 110:1107–1112
Stepanova AN, Robertson-Hoyt J, Yun J et al (2008) TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133:177–191
Tao Y, Ferrer JL, Ljung K et al (2008) Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133:164–176
Won C, Shen X, Mashiguchi K et al (2011) Conversion of tryptophan to indole-3-acetic acid by TRYPTOPHAN AMINOTRANSFERASES OF ARABIDOPSIS and YUCCAs in Arabidopsis. Proc Natl Acad Sci USA 108:18518–18523
Dai X, Mashiguchi K, Chen Q et al (2013) The biochemical mechanism of auxin biosynthesis by an Arabidopsis YUCCA flavin-containing monooxygenase. J Biol Chem 288:1448–1457
Mashiguchi K, Tanaka K, Sakai T et al (2011) The main auxin biosynthesis pathway in Arabidopsis. Proc Natl Acad Sci USA 108:18512–18517
Phillips KA, Skirpan AL, Liu X et al (2011) vanishing tassel2 encodes a grass-specific tryptophan aminotransferase required for vegetative and reproductive development in maize. Plant Cell 23:550–566
Stepanova AN, Yun J, Robles LM et al (2011) The Arabidopsis YUCCA1 flavin monooxygenase functions in the indole-3-pyruvic acid branch of auxin biosynthesis. Plant Cell 23:3961–3973
Zhao Y, Hull AK, Gupta NR et al (2002) Trp-dependent auxin biosynthesis in Arabidopsis: involvement of cytochrome P450 s CYP79B2 and CYP79B3. Genes Dev 16:3100–3112
Zhao Y, Christensen SK, Fankhauser C et al (2001) A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 291:306–309
Pedmale UV, Huang SS, Zander M et al (2016) Cryptochromes interact directly with PIFs to control plant growth in limiting blue light. Cell 164:233–245
Zheng Z, Guo Y, Novak O et al (2013) Coordination of auxin and ethylene biosynthesis by the aminotransferase VAS1. Nat Chem Biol 9:244–246
Nishizawa T, Aldrich CC, Sherman DH (2005) Molecular analysis of the rebeccamycin l-amino acid oxidase from Lechevalieria aerocolonigenes ATCC 39243. J Bacteriol 187:2084–2092
Onaka H, Taniguchi S, Igarashi Y et al (2003) Characterization of the biosynthetic gene cluster of rebeccamycin from Lechevalieria aerocolonigenes ATCC 39243. Biosci Biotechnol Biochem 67:127–138
Howard-Jones AR, Walsh CT (2005) Enzymatic generation of the chromopyrrolic acid scaffold of rebeccamycin by the tandem action of RebO and RebD. Biochemistry 44:15652–15663
Sugawara S, Hishiyama S, Jikumaru Y et al (2009) Biochemical analyses of indole-3-acetaldoxime-dependent auxin biosynthesis in Arabidopsis. Proc Natl Acad Sci USA 106:5430–5435
Hobbie L, Estelle M (1994) Genetic approaches to auxin action. Plant, Cell Environ 17:525–540
Yamada M, Greenham K, Prigge MJ et al (2009) The TRANSPORT INHIBITOR RESPONSE2 gene is required for auxin synthesis and diverse aspects of plant development. Plant Physiol 151:168–179
Radwanski ER, Last RL (1995) Tryptophan biosynthesis and metabolism: biochemical and molecular genetics. Plant Cell 7:921–934
Boerjan W, Cervera MT, Delarue M et al (1995) Superroot, a recessive mutation in Arabidopsis, confers auxin overproduction. Plant Cell 7:1405–1419
Delarue M, Prinsen E, Onckelen HV et al (1998) Sur2 mutations of Arabidopsis thaliana define a new locus involved in the control of auxin homeostasis. Plant J 14:603–611
He W, Brumos J, Li H et al (2011) A small-molecule screen identifies l-kynurenine as a competitive inhibitor of TAA1/TAR activity in ethylene-directed auxin biosynthesis and root growth in Arabidopsis. Plant Cell 23:3944–3960
Romano CP, Robson PR, Smith H et al (1995) Transgene-mediated auxin overproduction in Arabidopsis: hypocotyl elongation phenotype and interactions with the hy6-1 hypocotyl elongation and axr1 auxin-resistant mutants. Plant Mol Biol 27:1071–1083
Pacheco-Villalobos D, Sankar M, Ljung K et al (2013) Disturbed local auxin homeostasis enhances cellular anisotropy and reveals alternative wiring of auxin-ethylene crosstalk in Brachypodium distachyon seminal roots. PLoS Genet 9:e1003564
Acknowledgement
This work was supported by the NIH (R01GM114660 to YZ). ZZ and JC are supported by NIH (GM52413 to JC) and HHMI. JC is a HHMI investigator.
Author contributions
YZ and DB conceived the research. YG, XD, and YZ designed the experiments. YG and XD conducted the overexpression of RebO and analyzed the results. YK and HK did the auxin analysis. JC and ZZ conducted the shade avoidance assay. YZ wrote the paper.
Author information
Authors and Affiliations
Corresponding author
Additional information
Yangbin Gao and Xinhua Dai contributed equally to this work.
SPECIAL TOPIC: Plant Development and Reproduction.
About this article
Cite this article
Gao, Y., Dai, X., Zheng, Z. et al. Overexpression of the bacterial tryptophan oxidase RebO affects auxin biosynthesis and Arabidopsis development. Sci. Bull. 61, 859–867 (2016). https://doi.org/10.1007/s11434-016-1066-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11434-016-1066-2