Abstract
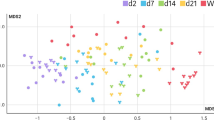
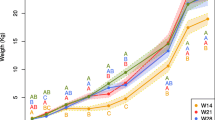
Gut microbiota play important roles in host health and diseases. Several factors, in particular antibiotics, affect the gut microbiota of pigs. Cross-fostering has been applied as a regular practice to equalize litter size, reduce pre-weaning mortality and increase body weight. However, the effect of cross-fostering on cecal microbiota is unclear. In this study, we fostered three Jinhua pigs to a Yorkshire sow. The fostered Jinhua piglets grew significantly faster than their biological siblings. To explore whether the cecal microbiota of piglets will alter during fostering, we characterized the cecal microbiota of all piglets by examining the V3 hypervariable region of 16S rDNA. We observed altered cecal microbiota in these piglets using the Illumina HiSeq 2000 platform, and this was accompanied with an increase in growth rate after fostering. The relative abundance of Bacteroidetes in the fostered Jinhua piglets was decreased compared with their biological siblings, although still higher in comparison with their new littermates. Beta-diversity analysis also showed that the cecal microbiota of the adopted Jinhua piglets differed from their biological siblings with a shift toward their step-siblings. Our data show that cecal microbiota of piglets were altered after cross-fostering while the growth rate increased.




Similar content being viewed by others
References
Meurens F, Summerfield A, Nauwynck H et al (2012) The pig: a model for human infectious diseases. Trends Microbiol 20:50–57
Hvistendahl M (2012) Pigs as stand-ins for microbiome studies. Science 336:1250
Delgado CL, Courbois C, Rosegrant MW (1998) Global food demand and the contribution of livestock as we enter the new millennium. International Food Policy Research Institute (IFPRI), Washington, DC
Straw BE, Burgi EJ, Dewey CE et al (1998) Effects of extensive crossfostering on performance of pigs on a farm. J Am Vet Med Assoc 212:855–856
Robert S, Martineau G (2001) Effects of repeated cross-fosterings on preweaning behavior and growth performance of piglets and on maternal behavior of sows. J Anim Sci 79:88–93
Rzasa A, Poznanski W, Akincza J et al (2002) The influence of primiparous sow litter standardization on their performance. Ann Anim Sci 2(Suppl):167–172
English PR, Smith WJ, MacLean A (1978) The sow-improving her efficiency. Farming Press, Ipswich
Heim G, Mellagi A, Bierhals T et al (2012) Effects of cross-fostering within 24 h after birth on pre-weaning behaviour, growth performance and survival rate of biological and adopted piglets. Livest Sci 150:121–127
Ley RE, Backhed F, Turnbaugh P et al (2005) Obesity alters gut microbial ecology. Proc Natl Acad Sci USA 102:11070–11075
Ley RE, Turnbaugh PJ, Klein S et al (2006) Microbial ecology: human gut microbes associated with obesity. Nature 444:1022–1023
Turnbaugh PJ, Ley RE, Mahowald MA et al (2006) An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444:1027–1031
Jumpertz R, Le DS, Turnbaugh PJ et al (2011) Energy-balance studies reveal associations between gut microbes, caloric load, and nutrient absorption in humans. Am J Clin Nutr 94:58–65
Round JL, Mazmanian SK (2009) The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol 9:313–323
Chung H, Pamp SJ, Hill JA et al (2012) Gut immune maturation depends on colonization with a host-specific microbiota. Cell 149:1578–1593
Diaz Heijtz R, Wang S, Anuar F et al (2011) Normal gut microbiota modulates brain development and behavior. Proc Natl Acad Sci USA 108:3047–3052
Marques TM, Wall R, Ross RP et al (2010) Programming infant gut microbiota: influence of dietary and environmental factors. Curr Opin Biotechnol 21:149–156
Spor A, Koren O, Ley R (2011) Unravelling the effects of the environment and host genotype on the gut microbiome. Nat Rev Microbiol 9:279–290
Lamendella R, Domingo JW, Ghosh S et al (2011) Comparative fecal metagenomics unveils unique functional capacity of the swine gut. BMC Microbiol 11:103
Kim HB, Borewicz K, White BA et al (2011) Longitudinal investigation of the age-related bacterial diversity in the feces of commercial pigs. Vet Microbiol 153:124–133
Looft T, Johnson TA, Allen HK et al (2012) In-feed antibiotic effects on the swine intestinal microbiome. Proc Natl Acad Sci USA 109:1691–1696
Kim HB, Borewicz K, White BA et al (2012) Microbial shifts in the swine distal gut in response to the treatment with antimicrobial growth promoter, tylosin. Proc Natl Acad Sci USA 109:15485–15490
Poole T, Suchodolski J, Callaway T et al (2013) The effect of chlortetracycline on faecal microbial populations in growing swine. J Glob Antimicrob Resist 3:171–174
Rettedal E, Vilain S, Lindblom S et al (2009) Alteration of the ileal microbiota of weanling piglets by the growth-promoting antibiotic chlortetracycline. Appl Environ Microbiol 75:5489–5495
Poroyko V, White JR, Wang M et al (2010) Gut microbial gene expression in mother-fed and formula-fed piglets. PLoS One 5:e12459
Pieper R, Janczyk P, Zeyner A et al (2008) Ecophysiology of the developing total bacterial and lactobacillus communities in the terminal small intestine of weaning piglets. Microb Ecol 56:474–483
Jacobson M, Hård af Segerstad C, Gunnarsson A et al (2003) Diarrhoea in the growing pig—a comparison of clinical, morphological and microbial findings between animals from good and poor performance herds. Res Vet Sci 74:163–169
Miao ZG, Wang LJ, Xu ZR et al (2009) Developmental changes of carcass composition, meat quality and organs in the Jinhua pig and Landrace. Animal 3:468–473
Yang F, Wang Q, Wang M et al (2012) Associations between gene polymorphisms in two crucial metabolic pathways and growth traits in pigs. Chin Sci Bull 57:2733–2740
Huse SM, Dethlefsen L, Huber JA et al (2008) Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing. PLoS Genetics 4:e1000255
Schloss PD, Westcott SL, Ryabin T et al (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541
Kozich JJ, Westcott SL, Baxter NT et al (2013) Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol 79:5112–5120
Chao A, Shen T-J (2003) Nonparametric estimation of Shannon’s index of diversity when there are unseen species in sample. Environ Ecol Stat 10:429–443
Segata N, Izard J, Waldron L et al (2011) Metagenomic biomarker discovery and explanation. Genome Biol 12:R60
Zou S, McLaren D, Hurley W (1992) Pig colostrum and milk composition: comparisons between Chinese Meishan and US breeds. Livest Product Sci 30:115–127
Ramanau A, Kluge H, Spilke J et al (2004) Supplementation of sows with L-carnitine during pregnancy and lactation improves growth of the piglets during the suckling period through increased milk production. J Nutr 134:86–92
Averette LA, Odle J, Monaco MH et al (1999) Dietary fat during pregnancy and lactation increases milk fat and insulin-like growth factor I concentrations and improves neonatal growth rates in swine. J Nutr 129:2123–2129
Gill SR, Pop M, Deboy RT et al (2006) Metagenomic analysis of the human distal gut microbiome. Science 312:1355–1359
Backhed F, Ding H, Wang T et al (2004) The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci USA 101:15718–15723
Backhed F, Manchester JK, Semenkovich CF et al (2007) Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc Natl Acad Sci USA 104:979–984
Martin FP, Dumas ME, Wang Y et al (2007) A top-down systems biology view of microbiome-mammalian metabolic interactions in a mouse model. Mol Syst Biol 3:112
Dominguez-Bello MG, Costello EK, Contreras M et al (2010) Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci USA 107:11971–11975
Turnbaugh PJ, Ridaura VK, Faith JJ et al (2009) The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med 1:6ra14
Wu GD, Chen J, Hoffmann C et al (2011) Linking long-term dietary patterns with gut microbial enterotypes. Science 334:105–108
Ley RE, Peterson DA, Gordon JI (2006) Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 124:837–848
Song SJ, Lauber C, Costello EK et al (2013) Cohabiting family members share microbiota with one another and with their dogs. eLife 2:e00458
Thompson CL, Wang B, Holmes AJ (2008) The immediate environment during postnatal development has long-term impact on gut community structure in pigs. ISME J 2:739–748
Zoetendal EG, Akkermans AD, Akkermans-van Vliet WM et al (2001) The host genotype affects the bacterial community in the human gastronintestinal tract. Microb Ecol Health Dis 13:129–134
Martín R, Langa S, Reviriego C et al (2003) Human milk is a source of lactic acid bacteria for the infant gut. J Pediatrics 143:754–758
Jimenez E, Delgado S, Maldonado A et al (2008) Staphylococcus epidermidis: a differential trait of the fecal microbiota of breast-fed infants. BMC Microbiol 8:143
Adlerberth I, Wold AE (2009) Establishment of the gut microbiota in Western infants. Acta Paediatr 98:229–238
O’Toole PW, Claesson MJ (2010) Gut microbiota: changes throughout the lifespan from infancy to elderly. Int Dairy J 20:281–291
Acknowledgments
We are grateful to Pinger Lou and Xiaoli Zhang for assistance with sample collections. We also thank Xingyong Ma for critically reading this manuscript. This work was supported by the National Special Foundation for Transgenic Species of China (2014ZX0800950B, 2011ZX08006-003), the Specialized Research Fund of Ministry of Agriculture of China (NYCYTX-009), the Fund of Fok Ying-Tung Education Foundation (141117), the Fund for Distinguished Young Scientists of Sichuan Province (2013JQ0013), the Program for Changjiang Scholars and Innovative Research Team in University (IRT13083) to Mingzhou Li and Xuewei Li, and the “100-Talent Program” in Sichuan Science Foundation for Youths of Sichuan Province to Ying Li.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Lingjin Xian, Ying Li and Zhi Jiang contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
11434_2014_532_MOESM1_ESM.xls
The electronic supplementary material is available online at csb.scichina.com and www.springerlink.com. The supporting materials are published as submitted, without typesetting or editing. The responsibility for scientific accuracy and content remains entirely with the authors (XLS 23 kb)
11434_2014_532_MOESM2_ESM.xls
The electronic supplementary material is available online at csb.scichina.com and www.springerlink.com. The supporting materials are published as submitted, without typesetting or editing. The responsibility for scientific accuracy and content remains entirely with the authors (XLS 26 kb)
11434_2014_532_MOESM3_ESM.xls
The electronic supplementary material is available online at csb.scichina.com and www.springerlink.com. The supporting materials are published as submitted, without typesetting or editing. The responsibility for scientific accuracy and content remains entirely with the authors (XLS 29 kb)
11434_2014_532_MOESM4_ESM.tif
The electronic supplementary material is available online at csb.scichina.com and www.springerlink.com. The supporting materials are published as submitted, without typesetting or editing. The responsibility for scientific accuracy and content remains entirely with the authors (TIFF 2784 kb)
About this article
Cite this article
Xian, L., Li, Y., Jiang, Z. et al. Alterations in cecal microbiota of Jinhua piglets fostered by a Yorkshire sow. Chin. Sci. Bull. 59, 4304–4311 (2014). https://doi.org/10.1007/s11434-014-0532-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11434-014-0532-y