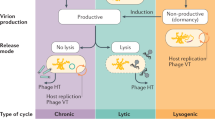
Abstract
Viruses are the most abundant biological entities in marine ecosystems. Most of them are phages that infect bacteria and archaea. Phages play important roles in causing the mortality of prokaryotic cells, structuring microbial communities, mediating horizontal gene transfer between different microbes, influencing the microbial food web process, and promoting biogeochemical cycles (such as C, N, etc.) in the ocean. Here we provided an overview of recent advances in research on the interactions between marine microorganisms and their phages, and suggest future research directions based on our understanding of the literature and our own work.
Article PDF
Similar content being viewed by others
Avoid common mistakes on your manuscript.
References
Jiao N Z. Marine Microbial Ecology (in Chinese). Beijing: Science Press, 2006. 272–303
Duckworth D H. History and basic properties of bacterial viruses. In: Goyal S M, Gerba C P, Bitton G, eds. Phage Ecology. New York: John Wiley & Sons, 1987. 1–44
Fuhrman J A. Marine viruses and their biogeochemical and ecological effects. Nature, 1999, 399: 541–548
Suttle C A. Marine viruses—Major players in the global ecosystem. Nat Rev Microbiol, 2007, 5: 801–812
Suttle C A. Viruses in the sea. Nature, 2005, 437: 356–361
Fuhrman J A, Suttle C A. Viruses in marine planktonic systems. Oceanography, 1993, 6: 51–63
Steward G F, Smith D C, Azam F. Abundance and production of bacteria and viruses in the Bering and Chukchi Seas. Mar Ecol-Prog Ser, 1996, 131: 287–300
Weinbauer M G, Hofle M G. Significance of viral lysis and flagellate grazing as factors controlling bacterioplankton production in a eutrophic lake. Appl Environ Microbiol, 1998, 64: 431–438
Suttle C A. The significance of viruses to mortality in aquatic microbial communities. Microb Ecol, 1994, 28: 237–243
Weinbauer M G. Ecology of prokaryotic viruses. FEMS Microbiol Rev, 2004, 28: 127–181
Danovaro R, Dell A, Corinaldesi C, et al. Major viral impact on the functioning of benthic deep-sea ecosystems. Nature, 2008, 454: 1084–1087
Wang F, Zheng T L, Hong H S. The important roles of marine virus in microbial food loop (in Chinese). Mar Sci, 1998, 4: 41–43
Wilhelm S W, Suttle C A. Viruses and Nutrient Cycles in the Sea. Bioscience, 1999, 49: 781–788
Wommack K E, Colwell R R. Virioplankton: Viruses in aquatic ecosystems. Microbiol Mol Biol Rev, 2000, 64: 69–114
Thingstad T F, Lignell R. Theoretical models for the control of bacterial growth rate, abundance, diversity and carbon demand. Aquat Microb Ecol, 1997, 13: 19–27
Zhang R, Weinbauer M G, Qian P Y. Viruses and flagellates sustain apparent richness and reduce biomass accumulation of bacterioplankton in coastal marine waters. Environ Microbiol, 2007, 9: 3008–3018
Allison G E, Klaenhammer T R. Phage resistance mechanisms in lactic acid bacteria. Inter Dairy J, 1998, 8: 207–226
Bohannan B J M, Kerr B, Jessup C M, et al. Trade-offs and coexistence in microbial microcosms. Antonie van Leeuwenhoek, 2002, 81: 107–115
Middelboe M, Holmfeldt K, Riemann L, et al. Bacteriophages drive strain diversification in a marine Flavobacterium: Implications for phage resistance and physiological properties. Environ Microbiol, 2009, 11: 1971–1982
Moebus K, Nattkemper H. Bacteriophage sensitivity patterns among bacteria isolated from marine waters. Helgoland Mar Res, 1981, 34: 375–385
Echols H. Developmental pathways for the temperate phage: Lysis vs lysogeny. Ann Rev Genet, 1976, 6: 157–190
Hewson I, O’Neil J M, Fuhrman J A, et al. Virus-like particle distribution and abundance in sediments and overlying waters along eutrophication gradients in two subtropical estuaries. Limnol Oceanogr, 2001, 46: 1734–1746
Lenski R E. Dynamics of interactions between bacteria and virulent bacteriophage. Adv Microb Ecol, 1988, 10: 1–44
Weinbauer M G, Suttle C A. Lysogeny and prophage induction in coastal and offshore bacterial communities. Aquat Microb Ecol, 1999, 18: 217–225
Jiang S C, Paul J H. Occurrence of lysogenic bacteria in marine microbial communities as determined by prophage induction. Mar Ecol Prog Ser, 1996, 142: 27–38
Jiang S C, Paul J H. Seasonal and diel abundance of viruses and occurrence of lysogeny/bacteriocinogeny in the marine environment. Mar Ecol Prog Ser, 1994, 104: 163–172
Jiang S C, Paul J H. Significance of lysogeny in the marine environments: studies with isolates and a model of lysogenic phage production. Microb Ecol, 1998, 35: 235–243
Wilhelm S W, Weinbauer M G, Suttle C A, et al. The role of sunlight in the removal and repair of viruses in the sea. Limnol Oceanogr, 1998, 43: 586–592
Weinbauer M G, Brettar I, Höfle M. Lysogeny and virus-induced mortality of bacterioplankton in surface, deep, and anoxic waters. Limnol Oceanogr, 2003, 48: 1457–1465
Ripp S, Miller R V. The role of pseudolysogeny in bacteriophagehost interactions in a natural freshwater environment. Microbiology, 1997, 143: 2065–2070
Ripp S, Miller R. Dynamics of the pseudolysogenic response in slowly growing cells of Pseudomonas aeruginosa. Microbiology, 1998, 144: 2225–2232
Moebus K. Marine bacteriophage reproduction under nutrient-limited growth of host bacteria. 2. Investigations with phage-host system [H3:H3/1]. Mar Ecol Prog Ser, 1996, 144: 13–22
Abedon S T. Selection for bacteriophage latent period length by bacterial density: A theoretical examination. Microb Ecol, 1989, 18: 79–88
Abedon S T, Herschler T D, Stopar D. Bacteriophage latent-period evolution as a response to resource availability. Appl Environ Microbiol, 2001, 67: 4233–4241
Rohwer F, Prangishvili D, Lindell D. Roles of viruses in the environment. Environ Microbiol, 2009, 11: 2771–2774
Davison J. Genetic exchange between bacteria in the environment. Plasmid, 1999, 42: 73–91
Dutta C, Pan A. Horizontal gene transfer and bacterial diversity. J Bios, 2002, 27: 27–33
Stotzky G. Gene transfer among bacteria in soil. In: Gene Transfer in the Environment. New York: McGraw-Hill, 1989. 165–222
Schicklmaier P, Schmieger H. Frequency of generalized transducing phages in natural isolates of the Salmonella typhimurium complex. Appl Environ Microbiol, 1995, 61: 1637–1640
Jiang S C, Paul J H. Gene transfer by transduction in the marine environment. Appl Environ Microbiol, 1998, 64: 2780–2787
Jain R, Rivera M C, Moore J E, et al. Horizontal Gene Transfer Accelerates Genome Innovation and Evolution. Mol Biol Evol, 2003, 20: 1598–1602
Lawrence J G. Gene transfer, speciation, and the evolution of bacterial genomes. Curr Opin Microbiol, 1999, 2: 519–523
Gogarten J P, Townsend J P. Horizontal gene transfer, genome innovation and evolution. Nat Rev Microbiol, 2005, 3: 679–687
Rohwer F, Thurber R V. Viruses manipulate the marine environment. Nature, 2009, 459: 207–212
Lindell D, Jaffe J D, Coleman M L, et al. Genome-wide expression dynamics of a marine virus and host reveal features of co-evolution. Nature, 2007, 449: 83–86
Lindell D, Sullivan M B, Johnson Z I, et al. Transfer of photosynthesis genes to and from Prochlorococcus viruses. Proc Nat Acad Sci USA, 2000, 101: 11013–11018
Lindell D, Jaffe J D, Johnson Z I, et al. Photosynthesis genes in marine viruses yield proteins during host infection. Nature, 2005, 438: 86–89
Nakayama K, Takashima K, Ishihara H, et al. The R-type pyocin of Pseudomonas aeruginosa is related to P2 phage, and the F-type is related to lambda phage. Mol Microbiol, 2000, 38: 213–231
Barrangou R, Fremaux C, Deveau H, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science, 2007, 315: 1709–1712
Cui Y J, Li Y J, Yan Y F, et al. Clustered regularly interspaced short palindromic repeats: Structure, function and application (in Chinese). Acta Microbiol Sin, 2008, 48: 1549–1555
Chibani-Chennoufi S, Bruttin A, Dillmann M L, et al. Phage-host interaction: An ecological perspective. Bacteriol, 2004, 186: 3677–3686
Huang C X, Zhang Y Y, Jiao N Z. Phage Resistance of a Marine Bacterium, Roseobacter denitrificans OCh114, as Revealed by Comparative Proteomics. Curr Microbiol, 2010, 61: 141–147
Hill C. Bacteriophage and bacteriophage resistance in lactic acid bacteria. FEMS Microbiol Rev, 1993, 12: 87–108
Forde A, Fitzgerald G F. Bacteriophage defence systems in lactic acid bacteria. Antonie van Leeuwenhoek, 1999, 76: 89–113
Bolotin A, Quinquis B, Sorokin A, et al. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiol, 2005, 151: 2551–2561
Mojica F J M, Díez-Villaseñor C, García-Martínez J, et al. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol, 2005, 60: 174–182
Pourcel C, Salvignol G, Vergnaud G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiol, 2005, 151: 653–663
Deveau H, Barrangou R, Garneau J E, et al. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J Bacteriol, 2008, 190: 1390–1400
Zhang Y Y, Jiao N Z, Colquhoun D R, et al. Protein modifications related to phage resistance in a marine roseobacter. Aquat Microb Ecol, 2009, 55: 203–207
Author information
Authors and Affiliations
Corresponding authors
Additional information
This article is published with open access at Springerlink.com
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Zhang, Y., Huang, C., Yang, J. et al. Interactions between marine microorganisms and their phages. Chin. Sci. Bull. 56, 1770–1777 (2011). https://doi.org/10.1007/s11434-011-4503-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11434-011-4503-2