Abstract
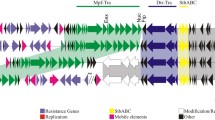
Based on three distinct traits of genomic islands, a novel approach was developed to search for and determine genomic islands in special strains. Two genomic islands in Pseudomonas aeruginosa PAO1 and 7 genomic islands in Pseudomonas aeruginosa PA14 were defined with this method. Among the 9 genomic islands, 4 islands had been characterized before, while the other 5 islands were initially determined. The insert sites of 6 genomic islands are tRNA sequences, direct repeats of PA14GI-3 are relative to tRNALeu, and direct repeats of PA14GI-2 are at the 3′ end of bifunctional GMP synthase/glutamine amidotransferase. Only direct repeats of PA14GI-4 are not clear. Among the 5 newly-found genomic islands, it was supposed that PA14GI-2 is a genomic island related to Hg2+ uptake, PA14GI-3 is a secretory activity genomic island, PA14GI-6 is a pathogenicity island, and functions of PA14GI-1 and PA14GI-5 are not clear. Finally, the tyrosine type integrases in PAO1GI-1, PA14GI-5 and PA14GI-7 were analyzed, and their binding and restriction sites were predicted.
Similar content being viewed by others
References
Dobrindt U, Hochhut B, Hentschel U, et al. Genomic islands in pathogenic and environmental microorganisms. Nat Rev Microbiol, 2004, 2: 414–424
Liò P, Vannucci M. Finding pathogenicity islands and gene transfer events in genome data. Bionformatics. 2000, 16: 932–940
Zhang R, Zhang C T. A systematic method to identify genomic islands and its applications in analyzing the genomes of Corynebacterium glutamicum and Vibrio vulnificus CMCP6 chromosome I. Bioinformatics, 2004, 20: 612–622
Mantri Y, Williams K P. Islander: A database of integrative islands in prokaryotic genomes, the associated integrases and their DNA site specificities. Nucleic Acids Res, 2004, 32: D55–D58
Langille M G, Hsiao W W, Brinkman F S. Evaluation of genomic island predictors using a comparative genomics approach. Bioinformatics, 2008, 9: 329
Ou H Y, He X, Harrison E M, et al. MobilomeFINDER: Web-based tools for in silico and experimental discovery of bacterial genomic islands. Nucleic Acids Res, 2007, 35: W97–W104
Langille M G, Brinkman F S. IslandViewer: An integrated interface for computational identification and visualization of genomic islands. Bioinformatics, 2009, 25: 664–665
Stover C K, Pham X Q, Erwin A L, et al. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature, 2000, 406: 959–964
Lee D G, Urbach J M, Wu G, et al. Genomic analysis reveals that Pseudomonas aeruginosa virulence is Combinatorial. Genome Biol, 2006, 7: R90
Kiewitz C, Larbig K, Klockgether J, et al. Monitoring genome evolution ex vivo: Reversible chromosomal integration of a 106-kb plasmid at two tRNA(Lys) gene loci in sequential Pseudomonas aeruginosa airway isolates. Microbiology, 2000, 146: 2365–2373
Webb J S, Lau M, Kjelleberg S. Bacteriophage and phenotypic variation in Pseudomonas aeruginosa biofilm development. J Bacteriol, 2004, 186: 8066–8073
He J, Baldini R L, Déziel E, et al. The broad host range pathogen Pseudomonas aeruginosa strain PA14 carries two pathogenicity islands harboring plant and animal virulence genes. Proc Natl Acad Sci USA, 2004, 101: 2530–2535
Mooij M J, Drenkard E, Llamas M A, et al. Characterization of the integrated filamentous phage Pf5 and its involvement in small-colony formation. Microbiology, 2007, 153: 1790–1798
van Passel M W, Luyf A C, van Kampen A H, et al. δρ-Web, an online tool to assess composition similarity of individual nucleic acid sequences. Bioinformatics, 2005, 21: 3053–3055
Ubeda C, Tormo M A, Cucarella C, et al. Sip, an integrase protein with excision, circularization and integration activities, defines a new family of mobile Staphylococcus aureus pathogenicity islands. Mol Microbiol, 2003, 49: 193–210
Pundhir S, Vijayvargiya H, Kumar A. PredictBias: A server for the identification of genomic and pathogenicity islands in prokaryotes. In Silico Biol, 2008, 8: 223–234
Lima-Mendez G, van Helden J, Toussaint A, et al. Prophinder: A computational tool for prophage prediction in prokaryotic genomes. Bioinformatics, 2008, 24: 863–865
Grindley N D, Whiteson K L, Rice P A. Mechanisms of site-specific recombination. Annu Rev Biochem, 2006, 75: 567–605
Author information
Authors and Affiliations
Corresponding author
Additional information
Supported by the National Natural Science Foundation of China (Grant Nos. 30821005 and 30870075), National Key Basic Research and Development Program of China (Grant No. 2009CB118906) and Shanghai Leading Academic Discipline Project (Grant No. B203)
About this article
Cite this article
Song, L., Zhang, X. Innovation for ascertaining genomic islands in PAO1 and PA14 of Pseudomonas aeruginosa . Chin. Sci. Bull. 54, 3991–3999 (2009). https://doi.org/10.1007/s11434-009-0598-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11434-009-0598-0