Abstract
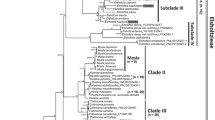
Araucariaceae is one of the most primitive families of the living conifers, and its phylogenetic relationships and divergence times are critically important issues. The DNA sequences of 8 genes, i.e., nuclear ribosomal 18S and 26S rRNA, chloroplast 16S rRNA, rbcL, matK and rps4, and mitochondrial coxI and atp1, obtained from this study and GenBank were used for constructing the molecular phylogenetic trees of Araucariaceae, indicating that the phylogenetic relationships among the three genera of this family should be ((Wollemia, Agathis), Araucaria). On the basis of the fossil calibrations of Wollemia and the two tribes Araucaria and Eutacta of the genus Araucaria, the divergence time of Araucariaceae was estimated to be (308 ± 53) million years ago, that is, the origin of the family was in the Late Carboniferous rather than Triassic as a traditional view. With the same gene combination, the divergence times of the genera Araucaria and Agathis were (246 ± 47) and (61 ± 15) Ma, respectively. Statistical analyses on the phylogenetic trees generated by using different genes and comparisons of the divergence times estimated by using those genes suggested that the chloroplast matK and rps4 genes are most suitable for investigating the phylogenetic relationships and divergence times of the family Araucariaceae.
Similar content being viewed by others
References
Farjon A. World Checklist and Bibliography of Conifers. 2nd. Richmond: Royal Botanic Gardens, 2001
Anonymous “Australia Hails a Prehistoric Pine” and “’Fossil Tree’ Reveals Full Splendour.” Nature, 1994, 372: 712, 719
Anderson I. Pine “dinosaur” Lurks in Gorge. New Scientist, 1994, 144: 5
Andrew S. Wollemia nobilis: A Living Fossil and Evolutionary Enigma. Institute for Creation Research, 2006, http://www.icr.org/article/2707/
Jones W G., K D Hill and J M Allen Wollemia nobilis, a new living Australian genus and species in the Araucariaceae. Telopea, 1995, 6: 173–176
Chambers T C, Drinnan A N, McLoughlin S. Some morphological features of wollemi pine (Wollemia nobilis: Araucariaceae) and their comparison to Cretaceous plant fossils. Int J Plant Sci, 1998, 159: 160–171
Setoguchi H, Ohsawa T A, Pintaud J C, et al. Phylogenetic relationships within Araucariaceae based on rbcL gene sequences. Am J Bot, 1998, 85: 1507–1516
Kershaw P, Wagstaff B. The southern conifer family Araucariaceae: History, status, and value for palaeoenvironmental reconstruction. Ann Rev Ecol Syst, 2001, 32: 397–414
Cantrill D J, Raine J I. Wairarapaia mildenhallii gen. et spec. nov., a new Araucarian cone related to Wollemia from the Cretaceous (Albian-Cenomanian) of New Zealand. Int J Plant Sci, 2006, 167: 1259–1269
Gilmore S, Hill K D. Relationships of the Wollemi Pine (Wollemia nobilis) and a molecular phylogeny of the Araucariaceae. Telopea, 1997, 7: 275–291
Stefanovi S, Jager M, Deutsch J, et al. Phylogenetic relationships of conifers inferred from partial 28S rRNA gene sequences. Am J Bot, 1998, 85: 688–697
Rydin C, Kallersjo M, Friis E M. Seed plant relationships and the systematic position of Gnetales based on nuclear and chloroplast DNA: Conflicting data, rooting problems and the monophyly of conifers. Int J Plant Sci, 2002, 163: 197–214
Chaw S M, Parkinson C L, Cheng Y, et al. Seed plant phylogeny inferred from all three-plant genomes: Monophyly of extant gymnosperms and origin of Gnetales from conifers. Proc Natl Acad Sci USA, 2000, 97: 4086–4091
Bowe M, Coat G, Claude W D. Phylogeny of seed plants based on all three genomic compartments: extant gymnosperms are monophyletic and Gnetales closest relatives are conifer. Proc Natl Acad Sci USA, 2000, 97: 4092–4097
Hart J A. A cladistic analysis of conifers: Preliminary results. J Arnold Arboretum, 1987, 68: 269–307
Price R A, Thomas J, Strauss S, et al. Familial relationships of the conifers from rbcL sequence data. Am J Bot, 1993, 80: 172
Miller C N. The origin of modern conifer families. In: Beck C B, ed. Origin and Evolution of Gymnosperms. New York: Columbia University Press, 1988. 448–486
Nixon K C, Crepet W L, Stevenson D, et al. A reevaluation of seed plant phylogeny. Ann Missouri Bot Gard, 1994, 81: 484–533
Doyle J J, Doyle J L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull, 1987, 19: 11–15
Rydin C, Pedersen K R, Friis E M. On the evolutionary history of Ephedra: Cretaceous fossils and extant molecules. Proc Natl Acad Sci USA, 2004, 101: 16571–16576
Ickert-Bond S M, Wojciechowski M E. Phylogenetic relationships in Ephedra (Gnetales): Evidence from nuclear and chloroplast DNA sequence data. Syst Bot, 2004, 29: 834–849
Gernano J, Klein A S. Species-specific nuclear and chloroplast single nucleotide polymorphisms to distinguish Picea glauca, P. mariana and P. rubens. Theor Appl Genet, 1999, 99: 37–49
Thompson J D, Gibson T J, Plewniak F, et al. The ClustaX windows interface: Flexible strategies for multiple sequences alignment aided by quality analysis tools. Nucleic Acids Res, 1997, 24: 4876–4882
Posada D, Crandall K A. Modeltest: Testing the model of DNA substitution. Bioinformatics, 1998, 14: 817–818
Guindon S, Lethiec F, Duroux P, et al. PHYML Online — a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res, 2005, 33(Suppl): W557–W559
Swofford D L. PAUP*: Phylogenetic anlysis using parsimony (* and other methods), Version 4. Sinauer Associates, Sunderland, Massachusetts, 2003
Shimodaira H, Hasegawa M. CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinformatics, 2001, 17: 1246–1247
Hill R S, Brodribb T J. Southern conifers in time and space. Austr J Bot, 1999, 47: 639–696
Yang Z. PAML 4: A program package for phylogenetic analysis by maximum likelihood. Mol Biol Evol, 2007, 24: 1586–1591
de Jersey N. Triassic spores and pollen grains from the Clematis Sanstone. Geol Surv Queensland Austr Publ, 1968, 338(Palaeont): 14: 1–44
Bamford M K, Philippe M. Jurassic-Early Cretaceous Gondwanan homoxylous woods: A nomenclatural revision of the genera with taxonomic notes. Rev Palaeobot Palynol, 2001, 113: 287–297
Pires E F, Guerra-Sommer M. Sommerxylon spiralosus from the Upper Triassic in southernmost Parana Basin (Brazil): A new taxon with taxacean affinity. Anais Acad Bras Cien, 2004, 76: 595–609
Author information
Authors and Affiliations
Corresponding author
Additional information
Supported by the National Infrastructure of Natural Resources for Science and Technology (Grant No. 2005DKA21403), MOST Basic Science and Technology (Grant No. 2007FY110100), Shanghai Leading Academic Discipline Project (Grant No. B111), Shanghai Science and Technology Committee (Grant No. 07XD14025), the Graduate Students Innovation Foundation of Fudan University (Grant No. EYH1322098) and National Science Fund for Fostering Talents in Basic Science (Grant No. J0630643)
About this article
Cite this article
Liu, N., Zhu, Y., Wei, Z. et al. Phylogenetic relationships and divergence times of the family Araucariaceae based on the DNA sequences of eight genes. Chin. Sci. Bull. 54, 2648–2655 (2009). https://doi.org/10.1007/s11434-009-0373-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11434-009-0373-2