Abstract
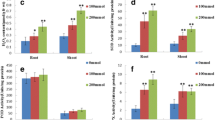
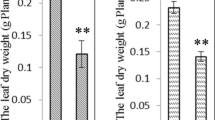
Salinity is one of the most severe environmental factors that may impair crop productivity. A proteomic study based on two-dimensional gel electrophoresis is performed in order to analyze the long-term salinity stress response of Thellungiella halophila, an Arabidopsis-related halophyte. Four-week-old seedlings are exposed to long-term salinity treatment. The total crude proteins are extracted from leaf blades, separated by 2-DE, stained with Coomassie Brilliant Blue, and differentially displayed spots are identified by MALDI-TOF MS or QTOF MS/MS. Among 900 protein spots reproducibly detected on each gel, 30 spots exhibit significant change and some of them are identified. The identified proteins include not only some previously characterized stress-responsive proteins such as TIR-NBS-LRR class disease resistance protein, ferritin-1, and pathogenesis-related protein 5, but also some proteins related to energy pathway, metabolism, RNA processing and protein degradation, as well as proteins with unknown functions. The possible functions of these proteins in salinity tolerance of T. halophila are discussed and it is suggested that the long-term salinity tolerance of T. halophila is achieved, at least partly, by enhancing defense system, adjusting energy and metabolic pathway and maintaining RNA structure.
Similar content being viewed by others
References
Flowers T J, Yeo A R. Breeding for salinity resistance in crop plants: Where next? Aust J Plant Physiol, 1995, 22(6): 875–884
Bressan R A, Zhang C, Zhang H, et al. Learning from the Arabidopsis experience. The next gene search paradigm. Plant Physiol, 2001, 127(4): 1354–1360
Inan G, Zhang Q, Li P, et al. Salt cress. A halophyte and cryophyte Arabidopsis relative model system and its applicability to molecular genetic analyses of growth and development of extremophiles. Plant Physiol, 2004, 135(3): 1718–1737
Wang Z, Li P, Fredricksen M, et al. Expressed sequence tags from Thellungiella halophila, a new model to study plant salt-tolerance. Plant Sci, 2004, 166(3): 609–616
Wong C E, Li Y, Whitty B R, et al. Expressed sequence tags from the Yukon ecotype of Thellungiella reveal that gene expression in response to cold, drought and salinity shows little overlap. Plant Mol Biol, 2005, 58(4): 561–574
Gong Q, Li P, Ma S, et al. Salinity stress adaptation competence in the extremophile Thellungiella halophila in comparison with its relative Arabidopsis thaliana. Plant J, 2005, 44(5): 826–839
Li P, Mane S P, Sioson A A. et al. Effects of chronic ozone exposure on gene expression in Arabidopsis thaliana ecotypes and in Thellungiella halophila. Plant Cell Environ, 2005, 29(5): 854–868
Taji T, Seki M, Satou M, et al. Comparative genomics in salt tolerance between Arabidopsis and Arabidopsis-related halophyte salt cress using Arabidopsis microarray. Plant Physiol, 2004, 135(3): 1697–1709
Wong C E, Li Y, Labbe A, et al. Transcriptional profiling implicates novel interactions between abiotic stress and hormonal responses in Thellungiella, a close relative of Arabidopsis. Plant Physiol, 2006, 140(4): 1437–1450
Volkov V, Amtmann A. Thellungiella halophila, a salt-tolerant relative of Arabidopsis thaliana, has specific root ion-channel features. Plant J, 2006, 48(3): 342–353
Pradet-Balade B, Boulme F, Beug H, et al. Translation control: bridging the gap between genomics and proteomics? Trends Biochem Sci, 2001, 26(4): 225–229
Zivy M, de Vienne D. Proteomics: A link between genomics, genetics and physiology. Plant Mol Biol, 2000, 44(5): 575–580
Kim D W, Rakwal R, Agrawal G K, et al. A hydroponic rice seedling culture model system for investigating proteome of salt stress in rice leaf. Electrophoresis, 2005, 26(23): 4521–4539
Askari H, Edqvist J, Hajheidari M, et al. Effects of salinity levels on proteome of Suaeda aegyptiaca leaves. Proteomics, 2006, 6(8): 2542–2554
Yan S, Tang Z, Su W, et al. Proteomic analysis of salt stress-responsive proteins in rice root. Proteomics, 2005, 5(1): 235–244
Jiang Y, Yang B, Harris N S, et al. Comparative proteomic analysis of NaCl stress-responsive proteins in Arabidopsis roots. J Exp Bot, 2007, 58(13): 3591–3607
Taylor N L, Heazlewood J L, Day D A, et al. Differential impact of environmental stresses on the pea mitochondrial proteome. Mol Cell Proteomics, 2005, 4(8): 1122–1133
Bray E A. Classification of genes differentially expressed during water deficit stress in Arabidopsis thaliana: An analysis using microarray and differential expression data. Ann Bot (Lond), 2002, 89(7): 803–811
Foyer C H, Lelandais M, Kunert K J. Photooxidative stress in plants. Physiol Plant, 1994, 92(4): 696–717
Deak M, Horvath G V, Davletova S, et al. Plants ectopically expressing the iron-binding protein, ferritin, are tolerant to oxidative damage and pathogens. Nat Biotechnol, 1999, 17(2): 192–196
Murgia I, Briat J F, Tarantino D. Plant ferritin accumulates in response to photoinhibition but its ectopic overexpression does not protect against photoinhibition. Plant Physiol Biochem, 2001, 39(9): 797–805
Seo P J, Lee A K, Xiang F, et al. Molecular and functional profiling of Arabidopsis pathogenesis-related genes: Insights into their roles in salt response of seed germination. Plant Cell Physiol, 2008, 49(3): 334–344
Amme S, Matros A, Schlesier B, et al. Proteome analysis of cold stress response in Arabidopsis thaliana using DIGE-technology. J Exp Bot, 2006, 57(7): 1537–1546
Yan S, Zhang Q, Tang Z, et al. Comparative proteomic analysis provides new insights into chilling stress responses in rice. Mol Cell Proteomics, 2006, 5(3): 484–496
Imin N, Kerim T, Rolfe B G, et al. Effect of early cold stress on the maturation of rice anthers. Proteomics, 2004, 4(7): 1873–1882
Ndimba B K, Chivasa S, Simon W J, et al. Identification of Arabidopsis salt and osmotic stress responsive proteins using two-dimensional difference gel electrophoresis and mass spectrometry. Proteomics, 2005, 5(16): 4185–4196
Kav N N V, Srivastava S, Goonewardene L, et al. Proteome-level changes in the roots of Pisum sativum in response to salinity. Ann Appl Biol, 2004, 145(2): 217–230
Russell D A, Sachs M M. Differential expression and sequence analysis of the maize glyceraldehyde-3-phosphate dehydrogenase gene family. Plant Cell, 1989, 1(8): 793–803
Baek D, Jin Y, Jeong J C, et al. Suppression of reactive oxygen species by glyceraldehyde-3-phosphate dehydrogenase. Phytochemistry, 2008, 69(2): 333–338
Dastoor Z, Dreyer J L. Potential role of nuclear translocation of glyceraldehyde-3-phosphate dehydrogenase in apoptosis and oxidative stress. J Cell Sci, 2001, 114(9): 1643–1653
Pilon-Smits E A, Hwang S, Mel Lytle C, et al. Overexpression of ATP sulfurylase in Indian mustard leads to increased selenate uptake, reduction, and tolerance. Plant Physiol, 1999, 119(1): 123–132
Alba M M, Pages M. Plant proteins containing the RNA-recognition motif. Trends Plant Sci, 1998, 3(6): 15–21
Mousavi A, Hotta Y. Glycine-rich proteins: A class of novel proteins. Appl Biochem Biotechnol, 2005, 120(3): 169–174
Sachetto-Martins G, Franco L O, de Oliveira D E. Plant glycine-rich proteins: A family or just proteins with a common motif? Biochim Biophys Acta, 2000, 1492(1): 1–14
Kim J S, Park S J, Kwak K J, et al. Cold shock domain proteins and glycine-rich RNA-binding proteins from Arabidopsis thaliana can promote the cold adaptation. Nucleic Acid Res, 2007, 35(2): 506–516
Mauro S, Dainese P, Lannoye R, et al. Cold-resistant and cold-sensitive maize lines differ in the phosphorylation of the photosystem II subunit, CP29. Plant Physiol, 1997, 115(1): 171–180
Yang P, Fu H, Walker J, et al. Purification of the Arabidopsis 26 S proteasome biochemical and molecular analyses revealed the presence of multiple isoforms. J Biol Chem, 2004, 279(8): 6401–6413
Author information
Authors and Affiliations
Corresponding authors
Additional information
Supported by the National Basic Research and Development Program of China (Grant Nos. 2006CB100100, 2004CB117303), National Natural Science Foundation of China (Grants Nos. 30670203, 30570434), Beijing Municipal National Natural Science Foundation (Grant No. 5062021), CUN 985-3-3 and Open Fund of Key Laboratory of Cell Proliferation and Regulation Biology, Ministry of Education of China.
About this article
Cite this article
Gao, F., Zhou, Y., Huang, L. et al. Proteomic analysis of long-term salinity stress-responsive proteins in Thellungiella halophila leaves. Chin. Sci. Bull. 53, 3530–3537 (2008). https://doi.org/10.1007/s11434-008-0455-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11434-008-0455-6