Abstract
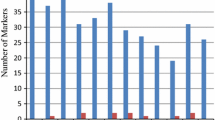
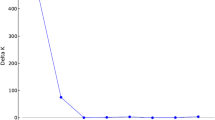
The pedigrees of three sequenced rice cultivars were analyzed to show that a majority of the genetic composition of ‘Nipponbare’ originates from japonica cultivars while the minority originates from indica cultivars. In contrast, ‘93-11’ is derived mainly from indica cultivars with a smaller contribution from japonica cultivars. All ancestors of ‘Guang lu ai 4’ appeared to be indica lines. A set of molecular markers (46 InDels and 53 SSRs) polymorphic between ‘Nipponbare’ and ‘93-11’ were examined in 46 typical indica and 47 typical japonica cultivars selected from 443 accessions according to Cheng’s index. All cultivars were divided into indica and japonica groups without overlapping when clustered by Cheng’s index, InDels and SSRs. Much higher InDel and SSR diversity between groups than within groups implies that the marker polymorphisms between ‘Nipponbare’ and ‘93-11’ represent a large proportion of inter-subspecific diversity. About 85% of indica cultivars and more than 90% of japonica cultivars were confirmed to have the same PCR banding patterns as ‘93-11’ and ‘Nipponbare’, respectively. Some polymorphic loci between ‘Nipponbare’ and ‘93-11’ cannot be validated in other indica and japonica cultivars, either as subspecies-specific but not predominant alleles, or alleles not specific between the two groups. It was concluded that molecular markers developed from sequence polymorphism between ‘Nipponbare’ and ‘93-11’ often represent inter-subspecific diversity, although some exceptions were sensitive to either particular marker loci or particular cultivars.
Similar content being viewed by others
References
Goff S A, Ricke D, Lan T H, et al. A draft sequence of the rice genome (Oryza sativa L ssp japonica). Science, 2002, 296: 92–100
Feng Q, Zhang Y, Hao P, et al. Sequence and analysis of rice chromosome 4. Nature, 2002, 420: 316–320
Sasaki T, Matsumoto T, Yamamoto K, et al. The genome sequence and structure of rice chromosome I. Nature, 2002, 420: 312–316
Yu J, Hu S, Wang J, et al. A draft sequence of the rice genome (Oryza sativa L ssp indica). Science, 2002, 296: 79–92
The Rice Chromosome 10 Sequencing Consortium. In-depth view of structure, activity, and evolution of rice chromosome 10. Science, 2003, 300: 1566–1569
Zhao Q, Zhang Y, Cheng Z, et al. A fine physical map of the rice chromosome-4. Genome Res, 2002, 12: 817–823
Han B, Xue Y B. Genome-wide intraspecific DNA-sequence variations in rice. Curr Opin in Plant Bio, 2003, 6: 134–138
Nasu S, Suzuki J, Ohta R, et al. Search for and analysis of single nucleotide polymorphisms SNPs) in rice (Oryza sativa, O. rufipogon) and establishment of SNP markers. DNA Res, 2002, 9: 163–171
Feltus F A, Wan J, Schulze S R, et al. An SNP resource for rice genetic and breeding based on subspecies indica and japonica genome alignments. Genome Res, 2004, 14: 1812–1819
Shen Y J, Jiang H, Jin J P, et al. Development of genome-wide DNA polymorphism database for map-based cloning of rice genes. Plant Physiol, 2004, 135: 1198–1205
Monna L, Ohta R, Masuda H, et al. Genome-wide searching of single-nucleotide polymorphisms among eight distantly and closely related rice cultivars (Oryza sativa L.) and a wild accession (Oryza rufipogon Griff.). DNA Res, 2006, 13: 43–51
Cheng K S. A statistical evaluation of the classification of rice cultivars into hsien and keng subspecies. Rice Genetics Newsletter, 1985, 2: 46–48
Murray M G, Thompson W F. Rapid isolation of high molecular weight plant DNA. Nuc Acid Res, 1980, 8: 4321–4325
Zhu Z F, Sun C Q, Li Z C, et al. Study on the classification of rice varieties with SSR molecular markers. Chin J Agri Biotech, 2001, 9: 58–61
Garris A J, Tai T H, Coburn J, et al. Genetic structure and diversity in Oryza sativa L. Genetics, 2005, 169: 1631–1638
Rohlf F J. NTSYS-pc, numerical taxonomy and multivariate analysis system, ver 2.02. Exeter Software, New York: Setauket, 1998
Yeh F C, Yang R C, Boyle T. POPGENE Version 1.31, Microsoft Windows-based Freeware for Population Genetic Analysis. Edmonton: University of Alberta, 1999
Kimura M, Crow J F. The number of alleles that can be maintained in a finite population. Genetics, 1964, 49: 725–738
Nei M. Analysis of gene diversity in subdivided populations. Proc Natl Acad of Sci USA, 1973, 70: 3321–3323
Lewontin R C. The apportionment of human diversity. Evo Bio, 1972, 6: 381–398
Schneider S, Roessi D, Excoffier L. Arlequin, a software for population genetics data analysis Ver 2.000. Genetics and Biometry Lab, Dept of Anthropology, University of Geneva, 2000
Rohlf F J. Algorithm 76, Hierarchical clustering using the minimum spanning tree. Comput J, 1973, 16: 93–95
Page R D M. TREEVIEW, An application to display phylogenetic trees on personal computers. Comput Appl Biosci, 1996, 12: 357–358
Glaszmann J C. Isozymes and classification of Asian rice. Theor Appl Genet, 1987, 74: 21–30
Li C, Zhang Y, Ying K, et al. Sequence variation of simple sequence repeats on chromosome 4 in two subspecies of the Asian cultivated rice. Theor Appl Genet, 2004, 108: 392–400
Zhang J W, Feng Q, Jin C Q, et al. Features of the expressed sequences revealed by a large-scale analysis of ESTs from a normalized cDNA library of the elite indica rice cultivar Minghui 63. Plant J, 2005, 42: 772–780
Khush G S. Origin, dispersal, cultivation and variation of rice. Plant Mol Bio, 1997, 35: 25–34
Oka H I. Origin of Cultivated Rice. Tokyo: Japan Scientific Societies Press, 1988
Second G. Origin of the genic diversity of cultivated rice (Oryza spp.), Study of the polymorphism scored at 40 isozyme loci. Jap J Genet, 1982, 57: 25–57
Kawase M, Kishimoto N, Tanaka T, et al. Intraspecific variation and genetic differentiation based on restriction fragment length polymorphism in Asian cultivated rice, Oryza sativa L. In: Rice Genetics II. Manila: International Rice Research Institute, 1992. 467–473
Zhang Q F, Maroof M A S, Lu T Y et al. Genetic diversity and differentiation of indica and japonica rice detected by RFLP analysis. Theor Appl Genet, 1992, 83: 495–499
Ni J, Colowit P M, Mackill D J. Evaluation of genetic diversity in rice subspecies using microsatellite markers. Crop Sci, 2002, 42: 601–607
Qian Q, He P, Zheng X, et al. Genetic analysis of morphological index and its related taxonomic traits for classification of indica/japonica rice. Sci China Ser C-Life Sci, 2000, 43(2): 113–119
Author information
Authors and Affiliations
Corresponding author
Additional information
Contributed equally to this paper
Supported by Shanghai Municipal Science and Technology Commission (Grant Nos. 03DJ14014 and 05DJ14008), Chinese Ministry of Agriculture (948 plan) (Grant Nos. 2001-101 and 2006-G-1) and Chinese Ministry of Science and Technology (Grant No. 2004CB117204)
About this article
Cite this article
Mei, H., Feng, F., Lu, B. et al. Experimental validation of inter-subspecific genetic diversity in rice represented by the differences between the DNA sequences of ‘Nipponbare’ and ‘93-11’. CHINESE SCI BULL 52, 1327–1337 (2007). https://doi.org/10.1007/s11434-007-0198-9
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/s11434-007-0198-9