Abstract
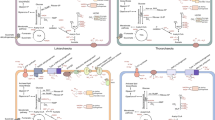
The hypothesis that eukaryotes originated from within the domain Archaea has been strongly supported by recent phylogenomic analyses placing Heimdallarchaeota-Wukongarchaeota branch from the Asgard superphylum as the closest known archaeal sister-group to eukaryotes. However, our understanding is still limited in terms of the relationship between eukaryotes and archaea, as well as the evolution and ecological functions of the Asgard archaea. Here, we describe three previously unknown phylum-level Asgard archaeal lineages, tentatively named Sigyn-, Freyr- and Njordarchaeota. Additional members in Wukongarchaeota and Baldrarchaeota from distinct environments are also reported here, further expanding their ecological roles and metabolic capacities. Comprehensive phylogenomic analyses further supported the origin of eukaryotes within Asgard archaea and a new lineage Njordarchaeota was supposed as the known closest branch with the eukaryotic nuclear host lineage. Metabolic reconstruction suggests that Njordarchaeota may have a heterotrophic lifestyle with capability of peptides and amino acids utilization, while Sigynarchaeota and Freyrarchaeota also have the potentials to fix inorganic carbon via the Wood-Ljungdahl pathway and degrade organic matters. Additionally, the Ack/Pta pathway for homoacetogenesis and de novo anaerobic cobalamin biosynthesis pathway were found in Freyrarchaeota and Wukongrarchaeota, respectively. Some previously unidentified eukaryotic signature proteins for intracellular membrane trafficking system, and the homologue of mu/sigma subunit of adaptor protein complex, were identified in Freyrarchaeota. This study expands the Asgard superphylum, sheds new light on the evolution of eukaryotes and improves our understanding of ecological functions of the Asgard archaea.
Similar content being viewed by others
References
Akıl, C., Tran, L.T., Orhant-Prioux, M., Baskaran, Y., Manser, E., Blanchoin, L., and Robinson, R.C. (2020). Insights into the evolution of regulated actin dynamics via characterization of primitive gelsolin/cofilin proteins from Asgard archaea. Proc Natl Acad Sci USA 117, 19904–19913.
Alneberg, J., Bjarnason, B.S., de Bruijn, I., Schirmer, M., Quick, J., Ijaz, U. Z., Lahti, L., Loman, N.J., Andersson, A.F., and Quince, C. (2014). Binning metagenomic contigs by coverage and composition. Nat Methods 11, 1144–1146.
Bolger, A.M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120.
Buchfink, B., Xie, C., and Huson, D.H. (2015). Fast and sensitive protein alignment using DIAMOND. Nat Methods 12, 59–60.
Bulzu, P.A., Andrei, A.Ş., Salcher, M.M., Mehrshad, M., Inoue, K., Kandori, H., Beja, O., Ghai, R., and Banciu, H.L. (2019). Casting light on Asgardarchaeota metabolism in a sunlit microoxic niche. Nat Microbiol 4, 1129–1137.
Cai, M., Liu, Y., Yin, X., Zhou, Z., Friedrich, M.W., Richter-Heitmann, T., Nimzyk, R., Kulkarni, A., Wang, X., Li, W., et al. (2020). Diverse Asgard archaea including the novel phylum Gerdarchaeota participate in organic matter degradation. Sci China Life Sci 63, 886–897.
Capella-Gutiérrez, S., Silla-Martínez, J.M., and Gabaldón, T. (2009). trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973.
Chaumeil, P.A., Mussig, A.J., Hugenholtz, P., and Parks, D.H. (2019). GTDB-Tk: a toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics 36, 1925–1927.
Dombrowski, N., Teske, A.P., and Baker, B.J. (2018). Expansive microbial metabolic versatility and biodiversity in dynamic Guaymas Basin hydrothermal sediments. Nat Commun 9, 1–3.
Doxey, A.C., Kurtz, D.A., Lynch, M.D.J., Sauder, L.A., and Neufeld, J.D. (2015). Aquatic metagenomes implicate Thaumarchaeota in global cobalamin production. ISME J 9, 461–471.
Embley, T.M., and Martin, W. (2006). Eukaryotic evolution, changes and challenges. Nature 440, 623–630.
Esser, C., Ahmadinejad, N., Wiegand, C., Rotte, C., Sebastiani, F., Gelius-Dietrich, G., Henze, K., Kretschmann, E., Richly, E., Leister, D., et al. (2004). A genome phylogeny for mitochondria among α-proteobacteria and a predominantly eubacterial ancestry of yeast nuclear genes. Mol Biol Evol 21, 1643–1660.
Fang, H., Li, D., Kang, J., Jiang, P., Sun, J., and Zhang, D. (2018). Metabolic engineering of Escherichia coli for de novo biosynthesis of vitamin B12. Nat Commun 9, 4917.
Feng, X., Wang, Y., Zubin, R., and Wang, F. (2019). Core metabolic features and hot origin of Bathyarchaeota. Engineering 5, 498–504.
Field, M.C., and Dacks, J.B. (2009). First and last ancestors: reconstructing evolution of the endomembrane system with ESCRTs, vesicle coat proteins, and nuclear pore complexes. Curr Opin Cell Biol 21, 4–13.
Fournier, G.P., and Gogarten, J.P. (2008). Evolution of acetoclastic methanogenesis in Methanosarcina via horizontal gene transfer from cellulolytic Clostridia. J Bacteriol 190, 1124–1127.
Hartman, H., and Fedorov, A. (2002). The origin of the eukaryotic cell: a genomic investigation. Proc Natl Acad Sci USA 99, 1420–1425.
He, Y., Li, M., Perumal, V., Feng, X., Fang, J., Xie, J., Sievert, S.M., and Wang, F. (2016). Genomic and enzymatic evidence for acetogenesis among multiple lineages of the archaeal phylum Bathyarchaeota widespread in marine sediments. Nat Microbiol 1, 16035.
Hirst, J., Barlow, L.D., Francisco, G.C., Sahlender, D.A., Seaman, M.N.J., Dacks, J.B., and Robinson, M.S. (2011). The fifth adaptor protein complex. PLoS Biol 9, e1001170.
Horton, P., Park, K.J., Obayashi, T., Fujita, N., Harada, H., Adams-Collier, C.J., and Nakai, K. (2007). WoLF PSORT: protein localization predictor. Nucleic Acids Res 35, W585–W587.
Huerta-Cepas, J., Szklarczyk, D., Heller, D., Hernández-Plaza, A., Forslund, S.K., Cook, H., Mende, D.R., Letunic, I., Rattei, T., Jensen, L.J., et al. (2019). eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res 47, D309–D314.
Hyatt, D., Chen, G.L., Locascio, P.F., Land, M.L., Larimer, F.W., and Hauser, L.J. (2010). Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinf 11, 119.
Imachi, H., Nobu, M.K., Nakahara, N., Morono, Y., Ogawara, M., Takaki, Y., Takano, Y., Uematsu, K., Ikuta, T., Ito, M., et al. (2020). Isolation of an archaeon at the prokaryote-eukaryote interface. Nature 577, 519–525.
Jay, Z.J., Beam, J.P., Dlakić, M., Rusch, D.B., Kozubal, M.A., and Inskeep, W.P. (2018). Marsarchaeota are an aerobic archaeal lineage abundant in geothermal iron oxide microbial mats. Nat Microbiol 3, 732–740.
Jones, P., Binns, D., Chang, H.Y., Fraser, M., Li, W., McAnulla, C., McWilliam, H., Maslen, J., Mitchell, A., Nuka, G., et al. (2014). InterProScan 5: genome-scale protein function classification. Bioinformatics 30, 1236–1240.
Kang, D.D., Li, F., Kirton, E., Thomas, A., Egan, R., An, H., and Wang, Z. (2019). MetaBAT 2: an adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ 7, e7359.
Katoh, K., and Standley, D.M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30, 772–780.
Lake, J.A., Henderson, E., Oakes, M., and Clark, M.W. (1984). Eocytes: a new ribosome structure indicates a kingdom with a close relationship to eukaryotes. Proc Natl Acad Sci USA 81, 3786–3790.
Lazar, C.S., Baker, B.J., Seitz, K., Hyde, A.S., Dick, G.J., Hinrichs, K.U., and Teske, A.P. (2016). Genomic evidence for distinct carbon substrate preferences and ecological niches of Bathyarchaeota in estuarine sediments. Environ Microbiol 18, 1200–1211.
Le, S.Q., and Gascuel, O. (2008). An improved general amino acid replacement matrix. Mol Biol Evol 25, 1307–1320.
Lee, I., Ouk Kim, Y., Park, S.C., and Chun, J. (2016). OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol 66, 1100–1103.
Leung, K.F., Dacks, J.B., and Field, M.C. (2008). Evolution of the multivesicular body ESCRT machinery; retention across the eukaryotic lineage. Traffic 9, 1698–1716.
Li, D., Liu, C.M., Luo, R., Sadakane, K., and Lam, T.W. (2015). MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31, 1674–1676.
Liu, Y., Makarova, K.S., Huang, W.C., Wolf, Y.I., Nikolskaya, A.N., Zhang, X., Cai, M., Zhang, C.J., Xu, W., Luo, Z., et al. (2021). Expanded diversity of Asgard archaea and their relationships with eukaryotes. Nature 593, 553–557.
Liu, Y., Zhou, Z., Pan, J., Baker, B.J., Gu, J.D., and Li, M. (2018). Comparative genomic inference suggests mixotrophic lifestyle for Thorarchaeota. ISME J 12, 1021–1031.
López-García, P., and Moreira, D. (2006). Selective forces for the origin of the eukaryotic nucleus. Bioessays 28, 525–533.
López-García, P., and Moreira, D. (2015). Open questions on the origin of eukaryotes. Trends Ecol Evol 30, 697–708.
López-García, P., and Moreira, D. (1999). Metabolic symbiosis at the origin of eukaryotes. Trends Biochem Sci 24, 88–93.
Luo, C., Rodriguez-R L.M., and Konstantinidis, K.T. (2014). MyTaxa: an advanced taxonomic classifier for genomic and metagenomic sequences. Nucleic Acids Res 42, e73.
MacLeod, F. S. Kindler, G., Lun Wong, H., Chen, R., and P. Burns, B. (2019). Asgard archaea: diversity, function, and evolutionary implications in a range of microbiomes. AIMS Microbiol 5, 48–61.
Makarova, K.S., Wolf, Y.I., and Koonin, E.V. (2015). Archaeal clusters of orthologous genes (arCOGs): an update and application for analysis of shared features between Thermococcales, Methanococcales, and Methanobacteriales. Life 5, 818–840.
Martin, W.F., Garg, S., and Zimorski, V. (2015). Endosymbiotic theories for eukaryote origin. Phil Trans R Soc B 370, 20140330.
Moreira, D., and López-García, P. (1998). Symbiosis between methanogenic archaea and δ-proteobacteria as the origin of eukaryotes: the syntrophic hypothesis. J Mol Evol 47, 517–530.
Moriya, Y., Itoh, M., Okuda, S., Yoshizawa, A.C., and Kanehisa, M. (2007). KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res 35, W182–W185.
Narrowe, A.B., Spang, A., Stairs, C.W., Caceres, E.F., Baker, B.J., Miller, C.S., and Ettema, T.J.G. (2018). Complex evolutionary history of translation Elongation Factor 2 and diphthamide biosynthesis in Archaea and parabasalids. Genome Biol Evol 10, 2380–2393.
Nguyen, L.T., Schmidt, H.A., von Haeseler, A., and Minh, B.Q. (2015). IQTREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32, 268–274.
Pan, J., Zhou, Z., Béjà, O., Cai, M., Yang, Y., Liu, Y., Gu, J.D., and Li, M. (2020). Genomic and transcriptomic evidence of light-sensing, porphyrin biosynthesis, Calvin-Benson-Bassham cycle, and urea production in Bathyarchaeota. Microbiome 8, 43.
Park, S.Y., and Guo, X. (2014). Adaptor protein complexes and intracellular transport. Biosci Rep 34, e00123.
Parks, D.H., Imelfort, M., Skennerton, C.T., Hugenholtz, P., and Tyson, G. W. (2015). CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25, 1043–1055.
Raiborg, C., and Stenmark, H. (2009). The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature 458, 445–452.
Rawlings, N.D., Barrett, A.J., and Finn, R. (2016). Twenty years of the MEROPS database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res 44, D343–D350.
Richter, K., Haslbeck, M., and Buchner, J. (2010). The heat shock response: life on the verge of death. Mol Cell 40, 253–266.
Rochette, N.C., Brochier-Armanet, C., and Gouy, M. (2014). Phylogenomic test of the hypotheses for the evolutionary origin of eukaryotes. Mol Biol Evol 31, 832–845.
Rother, M., and Metcalf, W.W. (2004). Anaerobic growth of Methanosarcina acetivorans C2A on carbon monoxide: an unusual way of life for a methanogenic archaeon. Proc Natl Acad Sci USA 101, 16929–16934.
Schulz, B.L., Stirnimann, C.U., Grimshaw, J.P.A., Brozzo, M.S., Fritsch, F., Mohorko, E., Capitani, G., Glockshuber, R., Grütter, M.G., and Aebi, M. (2009). Oxidoreductase activity of oligosaccharyltransferase subunits Ost3p and Ost6p defines site-specific glycosylation efficiency. Proc Natl Acad Sci USA 106, 11061–11066.
Seitz, K.W., Dombrowski, N., Eme, L., Spang, A., Lombard, J., Sieber, J. R., Teske, A.P., Ettema, T.J.G., and Baker, B.J. (2019). Asgard archaea capable of anaerobic hydrocarbon cycling. Nat Commun 10, 1822.
Seitz, K.W., Lazar, C.S., Hinrichs, K.U., Teske, A.P., and Baker, B.J. (2016). Genomic reconstruction of a novel, deeply branched sediment archaeal phylum with pathways for acetogenesis and sulfur reduction. ISME J 10, 1696–1705.
Søndergaard, D., Pedersen, C.N.S., and Greening, C. (2016). HydDB: a web tool for hydrogenase classification and analysis. Sci Rep 6, 34212.
Spang, A., Saw, J.H., Jørgensen, S.L., Zaremba-Niedzwiedzka, K., Martijn, J., Lind, A.E., van Eijk, R., Schleper, C., Guy, L., and Ettema, T.J.G. (2015). Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature 521, 173–179.
Spang, A., Stairs, C.W., Dombrowski, N., Eme, L., Lombard, J., Caceres, E.F., Greening, C., Baker, B.J., and Ettema, T.J.G. (2019). Proposal of the reverse flow model for the origin of the eukaryotic cell based on comparative analyses of Asgard archaeal metabolism. Nat Microbiol 4, 1138–1148.
Tan, J.Z.A., and Gleeson, P.A. (2019). Cargo sorting at the trans-Golgi network for shunting into specific transport routes: role of Arf small G proteins and adaptor complexes. Cells 8, 531.
Wang, Y., Wegener, G., Hou, J., Wang, F., and Xiao, X. (2019). Expanding anaerobic alkane metabolism in the domain of Archaea. Nat Microbiol 4, 595–602.
Williams, T.A., Cox, C.J., Foster, P.G., Szöllősi, G.J., and Embley, T.M. (2020). Phylogenomics provides robust support for a two-domains tree of life. Nat Ecol Evol 4, 138–147.
Williams, T.A., Foster, P.G., Cox, C.J., and Embley, T.M. (2013). An archaeal origin of eukaryotes supports only two primary domains of life. Nature 504, 231–236.
Woese, C.R., Kandler, O., and Wheelis, M.L. (1990). Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci USA 87, 4576–4579.
Wong, H.L., White Iii, R.A., Visscher, P.T., Charlesworth, J.C., Vázquez-Campos, X., and Burns, B.P. (2018). Disentangling the drivers of functional complexity at the metagenomic level in Shark Bay microbial mat microbiomes. ISME J 12, 2619–2639.
Wu, Y.W., Simmons, B.A., and Singer, S.W. (2016). MaxBin 2.0: an automated binning algorithm to recover genomes from multiple metagenomic datasets. Bioinformatics 32, 605–607.
Zaremba-Niedzwiedzka, K., Caceres, E.F., Saw, J.H., Bäckström, D., Juzokaite, L., Vancaester, E., Seitz, K.W., Anantharaman, K., Starnawski, P., Kjeldsen, K.U., et al. (2017). Asgard archaea illuminate the origin of eukaryotic cellular complexity. Nature 541, 353–358.
Zhang, H., Yohe, T., Huang, L., Entwistle, S., Wu, P., Yang, Z., Busk, P.K., Xu, Y., and Yin, Y. (2018). dbCAN2: a meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res 46, W95–W101.
Zhang, J.W., Dong, H.P., Hou, L.J., Liu, Y., Ou, Y.F., Zheng, Y.L., Han, P., Liang, X., Yin, G.Y., Wu, D.M., et al. (2021). Newly discovered Asgard archaea Hermodarchaeota potentially degrade alkanes and aromatics via alkyl/benzyl-succinate synthase and benzoyl-CoA pathway. ISME J 15, 1826–1843.
Zhang, R.Y., Zou, B., Yan, Y.W., Jeon, C.O., Li, M., Cai, M., and Quan, Z. X. (2020). Design of targeted primers based on 16S rRNA sequences in meta-transcriptomic datasets and identification of a novel taxonomic group in the Asgard archaea. BMC Microbiol 20, 25.
Zimmermann, L., Stephens, A., Nam, S.Z., Rau, D., Kübler, J., Lozajic, M., Gabler, F., Söding, J., Lupas, A.N., and Alva, V. (2018). A completely reimplemented MPI bioinformatics toolkit with a new HHpred server at its core. J Mol Biol 430, 2237–2243.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (91751205, 41921006, 41902313, 92051116), the China Ocean Mineral Resources R&D Association (COMRA) project (DY135-B2-12), the National Key Research and Development Project of China (2018YFC0310803), and the Senior User Project of RV KEXUE (KEXUE2019GZ06). We are grateful for Dr. Tom A. William for his suggestions regarding phylogenetic analysis. We thank Dr. Brett Baker for allowing us use their metagenomic data freely and these metagenomic data were produced by the US Department of Energy Joint Genome Institute http://www.jgi.doe.gov/ in collaboration with the user community. Thank Wan Liu for the genome submission to eLMSG (https://www.biosino.org/elmsg/index).
Author information
Authors and Affiliations
Corresponding author
Additional information
Compliance and ethics
The author(s) declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Xie, R., Wang, Y., Huang, D. et al. Expanding Asgard members in the domain of Archaea sheds new light on the origin of eukaryotes. Sci. China Life Sci. 65, 818–829 (2022). https://doi.org/10.1007/s11427-021-1969-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11427-021-1969-6