Abstract
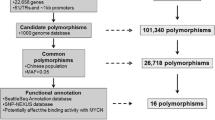
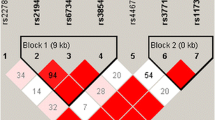
Neuroblastoma (NB) is one of the most common malignant tumors in children, with variable clinical behaviors and a 15% death rate of all malignancies in childhood. However, genetic susceptibility to sporadic NB in Han Chinese patients is largely unknown. To identify genetic risk factors for NB, we performed an association study on 357 NB patients and 738 control subjects among Han Chinese children. We focused on DEAD box 1 (DDX1), a putative RNA helicase, which is involved in NB carcinogenesis. The potential association of DDX1 polymorphisms with NB has not been discovered. Our results demonstrate that rs72780850 (NM_004939.2:c.-1555T>C) located in the DDX1 promoter region is significantly associated with higher expression of DDX1 transcript and increased NB risk (odds ratio=1.64, 95% confidence interval=1.03%–2.60%, P=0.004), especially in aggressive NB compared with ganglioneuroma and ganglioneuroblastoma in a dominant model (TC+CC vs. TT). Furthermore, the MYC-associated protein X (MAX) transcription factor showed stronger binding affinity to the DDX1 rs 72780850 CC allele compared with the TT allele, explaining the molecular mechanism of the increased NB risk caused by the rs72780850 polymorphism. Our results highlight the involvement of regulatory genetic variants of DDX1 in NB.
Similar content being viewed by others
References
Akter, K.A., Mansour, M.A., Hyodo, T., and Senga, T. (2017). FAM98A associates with DDX1-C14orf166-FAM98B in a novel complex involved in colorectal cancer progression. Int J Biochem Cell Biol 84, 1–13.
Bagatell, R., and Cohn, S.L. (2016). Genetic discoveries and treatment advances in neuroblastoma. Curr Opin Pediatr 28, 19–25.
Bown, N. (2001). Neuroblastoma tumour genetics: clinical and biological aspects. J Clin Pathol 54, 897–910.
Capasso, M., Devoto, M., Hou, C., Asgharzadeh, S., Glessner, J.T., Attiyeh, E.F., Mosse, Y.P., Kim, C., Diskin, S.J., Cole, K.A., et al. (2009). Common variations in BARD1 influence susceptibility to high-risk neuroblastoma. Nat Genet 41, 718–723.
Cascón, A., and Robledo, M. (2012). MAX and MYC: a heritable breakup. Cancer Res 72, 3119–3124.
Chou, C.F., Lin, W.J., Lin, C.C., Luber, C.A., Godbout, R., Mann, M., and Chen, C.Y. (2013). DEAD box protein DDX1 regulates cytoplasmic localization of KSRP. PLoS ONE 8, e73752.
Cimmino, F., Avitabile, M., Diskin, S.J., Vaksman, Z., Pignataro, P., Formicola, D., Cardinale, A., Testori, A., Koster, J., de Torres, C., et al. (2018). Fine mapping of 2q35 high-risk neuroblastoma locus reveals independent functional risk variants and suggests full-length BARD1 as tumor-suppressor. Int J Cancer 143, 2828–2837.
Diskin, S.J., Capasso, M., Schnepp, R.W., Cole, K.A., Attiyeh, E.F., Hou, C., Diamond, M., Carpenter, E.L., Winter, C., Lee, H., et al. (2012). Common variation at 6q16 within HACE1 and LIN28B influences susceptibility to neuroblastoma. Nat Genet 44, 1126–1130.
Ferrucci, F., Ciaccio, R., Monticelli, S., Pigini, P., di Giacomo, S., Purgato, S., Erriquez, D., Bernardoni, R., Norris, M., Haber, M., et al. (2018). MAX to MYCN intracellular ratio drives the aggressive phenotype and clinical outcome of high risk neuroblastoma. Biochim Biophys Acta 1861, 235–245.
Gabriel, S., Ziaugra, L., and Tabbaa, D. (2009). SNP genotyping using the Sequenom MassARRAY iPLEX platform. Curr Protoc Hum Genet 60.
Godbout, R., Li, L., Liu, R.Z., and Roy, K. (2007). Role of DEAD box 1 in retinoblastoma and neuroblastoma. Future Oncol 3, 575–587.
Godbout, R., and Squire, J. (1993). Amplification of a DEAD box protein gene in retinoblastoma cell lines. Proc Natl Acad Sci USA 90, 7578–7582.
Han, C., Liu, Y., Wan, G., Choi, H.J., Zhao, L., Ivan, C., He, X., Sood, A. K., Zhang, X., and Lu, X. (2014). The RNA-binding protein DDX1 promotes primary microRNA maturation and inhibits ovarian tumor progression. Cell Rep 8, 1447–1460.
He, J., Yang, T., Zhang, R., Zhu, J., Wang, F., Zou, Y., and Xia, H. (2016). Potentially functional polymorphisms in the LIN28B gene contribute to neuroblastoma susceptibility in Chinese children. J Cell Mol Med 20, 1534–1541.
He, J., Zou, Y., Wang, T., Zhang, R., Yang, T., Zhu, J., Wang, F., and Xia, H. (2017). Genetic variations of GWAS-identified genes and neuroblastoma susceptibility: a replication study in Southern Chinese children. Transl Oncol 10, 936–941.
Hua, R.X., Zhuo, Z., Ge, L., Zhu, J., Yuan, L., Chen, C., Liu, J., Cheng, J., Zhou, H., Zhang, J., et al. (2020). LIN28A gene polymorphisms modify neuroblastoma susceptibility: A four-centre case-control study. J Cell Mol Med 24, 1059–1066.
Li, L., Germain, D.R., Poon, H.Y., Hildebrandt, M.R., Monckton, E.A., McDonald, D., Hendzel, M.J., and Godbout, R. (2016). DEAD box 1 facilitates removal of RNA and homologous recombination at DNA double-strand breaks. Mol Cell Biol 36, 2794–2810.
Linder, P. (2006). Dead-box proteins: a family affair—Active and passive players in RNP-remodeling. Nucleic Acids Res 34, 4168–4180.
Lu, J., Chu, P., Wang, H., Jin, Y., Han, S., Han, W., Tai, J., Guo, Y., and Ni, X. (2015). Candidate gene association analysis of neuroblastoma in Chinese children strengthens the role of LMO1. PLoS ONE 10, e0127856.
Maris, J.M. (2010). Recent advances in neuroblastoma. N Engl J Med 362, 2202–2211.
Maris, J.M., Hogarty, M.D., Bagatell, R., and Cohn, S.L. (2007). Neuroblastoma. Lancet 369, 2106–2120.
Maris, J.M., Mosse, Y.P., Bradfield, J.P., Hou, C., Monni, S., Scott, R.H., Asgharzadeh, S., Attiyeh, E.F., Diskin, S.J., Laudenslager, M., et al. (2008). Chromosome 6p22 locus associated with clinically aggressive neuroblastoma. N Engl J Med 358, 2585–2593.
Nguyen, L.B., Diskin, S.J., Capasso, M., Wang, K., Diamond, M.A., Glessner, J., Kim, C., Attiyeh, E.F., Mosse, Y.P., Cole, K., et al. (2011). Phenotype restricted genome-wide association study using a genecentric approach identifies three low-risk neuroblastoma susceptibility loci. PLoS Genet 7, e1002026.
Oldridge, D.A., Wood, A.C., Weichert-Leahey, N., Crimmins, I., Sussman, R., Winter, C., McDaniel, L.D., Diamond, M., Hart, L.S., Zhu, S., et al. (2015). Genetic predisposition to neuroblastoma mediated by a LMO1 super-enhancer polymorphism. Nature 528, 418–421.
Ritenour, L.E., Randall, M.P., Bosse, K.R., and Diskin, S.J. (2018). Genetic susceptibility to neuroblastoma: current knowledge and future directions. Cell Tissue Res 372, 287–307.
Shimada, H., Ambros, I.M., Dehner, L.P., Hata, J., Joshi, V.V., and Roald, B. (1999). Terminology and morphologic criteria of neuroblastic tumors. Cancer 86, 349–363.
Shi, J., Yu, Y., Jin, Y., Lu, J., Zhang, J., Wang, H., Han, W., Chu, P., Tai, J., Chen, F., et al. (2019). Functional polymorphisms in BARD1 association with neuroblastoma in a regional Han Chinese population. J Cancer 10, 2153–2160.
Squire, J.A., Thorner, P.S., Weitzman, S., Maggi, J.D., Dirks, P., Doyle, J., Hale, M., and Godbout, R. (1995). Co-amplification of MYCN and a DEAD box gene (DDX1) in primary neuroblastoma. Oncogene 10, 1417–1422.
Tanaka, K., Okamoto, S., Ishikawa, Y., Tamura, H., and Hara, T. (2009). DDX1 is required for testicular tumorigenesis, partially through the transcriptional activation of 12p stem cell genes. Oncogene 28, 2142–2151.
Taunk, N.K., Goyal, S., Wu, H., Moran, M.S., Chen, S., and Haffty, B.G. (2012). DEAD box 1 (DDX1) expression predicts for local control and overall survival in early stage, node-negative breast cancer. Cancer 118, 888–898.
Vessey, J.P., Amadei, G., Burns, S.E., Kiebler, M.A., Kaplan, D.R., and Miller, F.D. (2012). An asymmetrically localized Staufen2-dependent RNA complex regulates maintenance of mammalian neural stem cells. Cell Stem Cell 11, 517–528.
Vo, K.T., Matthay, K.K., Neuhaus, J., London, W.B., Hero, B., Ambros, P. F., Nakagawara, A., Miniati, D., Wheeler, K., Pearson, A.D.J., et al. (2014). Clinical, biologic, and prognostic differences on the basis of primary tumor site in neuroblastoma: a report from the international neuroblastoma risk group project. J Clin Oncol 32, 3169–3176.
Wang, D., Hashimoto, H., Zhang, X., Barwick, B.G., Lonial, S., Boise, L. H., Vertino, P.M., and Cheng, X. (2017). MAX is an epigenetic sensor of 5-carboxylcytosine and is altered in multiple myeloma. Nucleic Acids Res 45, 2396–2407.
Wang, H., Tan, W., Hao, B., Miao, X., Zhou, G., He, F., and Lin, D. (2003). Substantial reduction in risk of lung adenocarcinoma associated with genetic polymorphism in CYP2A13, the most active cytochrome P450 for the metabolic activation of tobacco-specific carcinogen NNK. Cancer Res 63, 8057–8061.
Wang, K., Diskin, S.J., Zhang, H., Attiyeh, E.F., Winter, C., Hou, C., Schnepp, R.W., Diamond, M., Bosse, K., Mayes, P.A., et al. (2011). Integrative genomics identifies LMO1 as a neuroblastoma oncogene. Nature 469, 216–220.
Yu, A.L., Gilman, A.L., Ozkaynak, M.F., London, W.B., Kreissman, S.G., Chen, H.X., Smith, M., Anderson, B., Villablanca, J.G., Matthay, K.K., et al. (2010). Anti-GD2 antibody with GM-CSF, interleukin-2, and isotretinoin for neuroblastoma. N Engl J Med 363, 1324–1334.
Zhang, R., Zou, Y., Zhu, J., Zeng, X., Yang, T., Wang, F., He, J., and Xia, H. (2016). The association between GWAS-identified BARD1 gene SNPs and neuroblastoma susceptibility in a Southern Chinese population. Int J Med Sci 13, 133–138.
Zhang, Z., Zhang, R., Zhu, J., Wang, F., Yang, T., Zou, Y., He, J., and Xia, H. (2017). Common variations within HACE1 gene and neuroblastoma susceptibility in a Southern Chinese population. Onco Targets Ther Volume 10, 703–709.
Zhuo, Z.J., Zhang, R., Zhang, J., Zhu, J., Yang, T., Zou, Y., He, J., and Xia, H. (2018). Associations between lncRNA MEG3 polymorphisms and neuroblastoma risk in Chinese children. Aging 10, 481–491.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (81472369 and 81502144), Beijing Natural Science Foundation Program and Scientific Research Key Program of Beijing Municipal Commission of Education (KZ201810025034), Clinical Application Research Funds of Capital Beijing (Z171100001017051), Beihang University & Capital Medical University Advanced Innovation Center for Big Data-Based Precision Medicine Plan (BHME-201804 and BHME-201904), and The Special Fund of the Pediatric Medical Coordinated Development Center of Beijing Hospitals Authority (XTCX201806). The authors are grateful to the members of the Department of Surgery and Beijing Key Laboratory for Pediatric Diseases of Otolaryngology, Head and Neck Surgery of Beijing Children’s Hospital for invaluable technical assistance and advice.
Author information
Authors and Affiliations
Corresponding author
Additional information
Compliance and ethics
The author(s) declare that they have no conflict of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Jin, Y., Shi, J., Wang, H. et al. MYC-associated protein X binding with the variant rs72780850 in RNA helicase DEAD box 1 for susceptibility to neuroblastoma. Sci. China Life Sci. 64, 991–999 (2021). https://doi.org/10.1007/s11427-020-1784-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11427-020-1784-7