Abstract
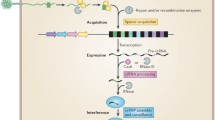
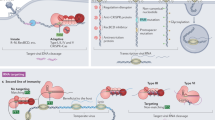
CRISPR-Cas systems provide the small RNA-based adaptive immunity to defend against invasive genetic elements in archaea and bacteria. Organisms of Sulfolobales, an order of thermophilic acidophiles belonging to the Crenarchaeotal Phylum, usually contain both type I and type III CRISPR-Cas systems. Two species, Saccharolobus solfataricus and Sulfolobus islandicus, have been important models for CRISPR study in archaea, and knowledge obtained from these studies has greatly expanded our understanding of molecular mechanisms of antiviral defense in all three steps: adaptation, expression and crRNA processing, and interference. Four subtypes of CRISPR-Cas systems are common in these organisms, including I-A, I-D, III-B, and III-D. These cas genes form functional modules, e.g., all genes required for adaptation and for interference in the I-A immune system are clustered together to form aCas and iCas modules. Genetic assays have been developed to study mechanisms of adaptation and interference by different CRISPR-Cas systems in these model archaea, and these methodologies are useful in demonstration of the protospacer-adjacent motif (PAM)-dependent DNA interference by I-A interference modules and multiple interference activities by III-B Cmr systems. Ribonucleoprotein effector complexes have been isolated for Sulfolobales III-B and III-D systems, and their biochemical characterization has greatly enriched the knowledge of molecular mechanisms of these novel antiviral immune responses.
Similar content being viewed by others
References
Albers, S.V., Jonuscheit, M., Dinkelaker, S., Urich, T., Kletzin, A., Tampe, R., Driessen, A.J.M., and Schleper, C. (2006). Production of recombinant and tagged proteins in the hyperthermophilic archaeon Sulfolobus solfataricus. Appl Environ Microbiol 72, 102–111.
Alkhnbashi, O.S., Costa, F., Shah, S.A., Garrett, R.A., Saunders, S.J., and Backofen, R. (2014). CRISPRstrand: predicting repeat orientations to determine the crRNA-encoding strand at CRISPR loci. Bioinformatics 30, i489–i496.
Athukoralage, J.S., McMahon, S.A., Zhang, C., Grüschow, S., Graham, S., Krupovic, M., Whitaker, R.J., Gloster, T.M., and White, M.F. (2020). An anti-CRISPR viral ring nuclease subverts type III CRISPR immunity. Nature 577, 572–575.
Athukoralage, J.S., Rouillon, C., Graham, S., Grüschow, S., and White, M. F. (2018). Ring nucleases deactivate type III CRISPR ribonucleases by degrading cyclic oligoadenylate. Nature 562, 277–280.
Barrangou, R., Fremaux, C., Deveau, H., Richards, M., Boyaval, P., Moineau, S., Romero, D.A., and Horvath, P. (2007). CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712.
Benda, C., Ebert, J., Scheltema, R.A., Schiller, H.B., Baumgärtner, M., Bonneau, F., Mann, M., and Conti, E. (2014). Structural model of a CRISPR RNA-silencing complex reveals the RNA-target cleavage activity in Cmr4. Mol Cell 56, 43–54.
Bhoobalan-Chitty, Y., Johansen, T.B., Di Cianni, N., and Peng, X. (2019). Inhibition of type III CRISPR-Cas immunity by an archaeal virus-encoded anti-CRISPR protein. Cell 179, 448–458.e11.
Bolotin, A., Quinquis, B., Sorokin, A., and Ehrlich, S.D. (2005). Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology 151, 2551–2561.
Bondy-Denomy, J., Pawluk, A., Maxwell, K.L., and Davidson, A.R. (2013). Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system. Nature 493, 429–432.
Brouns, S.J.J., Jore, M.M., Lundgren, M., Westra, E.R., Slijkhuis, R.J.H., Snijders, A.P.L., Dickman, M.J., Makarova, K.S., Koonin, E.V., and van der Oost, J. (2008). Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321, 960–964.
Carte, J., Wang, R., Li, H., Terns, R.M., and Terns, M.P. (2008). Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Genes Dev 22, 3489–3496.
Chen, K., Wang, Y., Zhang, R., Zhang, H., and Gao, C. (2019). CRISPR/Cas genome editing and precision plant breeding in agriculture. Annu Rev Plant Biol 70, 667–697.
Cheng, F., Gong, L., Zhao, D., Yang, H., Zhou, J., Li, M., and Xiang, H. (2017). Harnessing the native type I-B CRISPR-Cas for genome editing in a polyploid archaeon. J Genet Genomics 44, 541–548.
Cong, L., Ran, F.A., Cox, D., Lin, S., Barretto, R., Habib, N., Hsu, P.D., Wu, X., Jiang, W., Marraffini, L.A., et al. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823.
Contursi, P., Jensen, S., Aucelli, T., Rossi, M., Bartolucci, S., and She, Q. (2006). Characterization of the Sulfolobus host-SSV2 virus interaction. Extremophiles 10, 615–627.
Datsenko, K.A., Pougach, K., Tikhonov, A., Wanner, B.L., Severinov, K., and Semenova, E. (2012). Molecular memory of prior infections activates the CRISPR/Cas adaptive bacterial immunity system. Nat Commun 3, 945.
Deng, L., Garrett, R.A., Shah, S.A., Peng, X., and She, Q. (2013). A novel interference mechanism by a type IIIB CRISPR-Cmr module in Sulfolobus. Mol Microbiol 87, 1088–1099.
Deng, L., Kenchappa, C.S., Peng, X., She, Q., and Garrett, R.A. (2012). Modulation of CRISPR locus transcription by the repeat-binding protein Cbp1 in Sulfolobus. Nucleic Acids Res 40, 2470–2480.
Deng, L., Zhu, H., Chen, Z., Liang, Y.X., and She, Q. (2009). Unmarked gene deletion and host-vector system for the hyperthermophilic crenarchaeon Sulfolobus islandicus. Extremophiles 13, 735–746.
Deveau, H., Barrangou, R., Garneau, J.E., Labonte, J., Fremaux, C., Boyaval, P., Romero, D.A., Horvath, P., and Moineau, S. (2008). Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J Bacteriol 190, 1390–1400.
Elmore, J.R., Sheppard, N.F., Ramia, N., Deighan, T., Li, H., Terns, R.M., and Terns, M.P. (2016). Bipartite recognition of target RNAs activates DNA cleavage by the Type III-B CRISPR-Cas system. Genes Dev 30, 447–459.
Erdmann, S., and Garrett, R.A. (2012). Selective and hyperactive uptake of foreign DNA by adaptive immune systems of an archaeon via two distinct mechanisms. Mol Microbiol 85, 1044–1056.
Erdmann, S., Le Moine Bauer, S., and Garrett, R.A. (2014). Inter-viral conflicts that exploit host CRISPR immune systems of Sulfolobus. Mol Microbiol 91, 900–917.
Estrella, M.A., Kuo, F.T., and Bailey, S. (2016). RNA-activated DNA cleavage by the Type III-B CRISPR-Cas effector complex. Genes Dev 30, 460–470.
Forterre, P., and Prangishvili, D. (2009). The great billion-year war between ribosome- and capsid-encoding organisms (cells and viruses) as the major source of evolutionary novelties. Ann New York Acad Sci 1178, 65–77.
Foster, K., Kalter, J., Woodside, W., Terns, R.M., and Terns, M.P. (2019). The ribonuclease activity of Csm6 is required for anti-plasmid immunity by Type III-A CRISPR-Cas systems. RNA Biol 16, 449–460.
Fusco, S., Liguori, R., Limauro, D., Bartolucci, S., She, Q., and Contursi, P. (2015). Transcriptome analysis of Sulfolobus solfataricus infected with two related fuselloviruses reveals novel insights into the regulation of CRISPR-Cas system. Biochimie 118, 322–332.
Garrett, R.A., Shah, S.A., Erdmann, S., Liu, G., Mousaei, M., León-Sobrino, C., Peng, W., Gudbergsdottir, S., Deng, L., Vestergaard, G., et al. (2015). CRISPR-Cas adaptive immune systems of the Sulfolobales: unravelling their complexity and diversity. Life 5, 783–817.
Garrett, R.A., Shah, S.A., Vestergaard, G., Deng, L., Gudbergsdottir, S., Kenchappa, C.S., Erdmann, S., and She, Q. (2011a). CRISPR-based immune systems of the Sulfolobales: complexity and diversity. Biochem Soc Trans 39, 51–57.
Garrett, R.A., Vestergaard, G., and Shah, S.A. (2011b). Archaeal CRISPR-based immune systems: exchangeable functional modules. Trends Microbiol 19, 549–556.
Gudbergsdottir, S., Deng, L., Chen, Z., Jensen, J.V.K., Jensen, L.R., She, Q., and Garrett, R.A. (2011). Dynamic properties of the Sulfolobus CRISPR/Cas and CRISPR/Cmr systems when challenged with vector-borne viral and plasmid genes and protospacers. Mol Microbiol 79, 35–49.
Guo, L., Brügger, K., Liu, C., Shah, S.A., Zheng, H., Zhu, Y., Wang, S., Lillestøl, R.K., Chen, L., Frank, J., et al. (2011). Genome analyses of Icelandic strains of Sulfolobus islandicus, model organisms for genetic and virus-host interaction studies. J Bacteriol 193, 1672–1680.
Guo, T., Han, W., and She, Q. (2019a). Tolerance of Sulfolobus SMV1 virus to the immunity of I-A and III-B CRISPR-Cas systems in Sulfolobus islandicus. RNA Biol 16, 549–556.
Guo, T., Zheng, F., Zeng, Z., Yang, Y., Li, Q., She, Q., and Han, W. (2019b). Cmr3 regulates the suppression on cyclic oligoadenylate synthesis by tag complementarity in a Type III-B CRISPR-Cas system. RNA Biol 16, 1513–1520.
Haft, D.H., Selengut, J., Mongodin, E.F., and Nelson, K.E. (2005). A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput Biol 1, e60.
Hale, C.R., Zhao, P., Olson, S., Duff, M.O., Graveley, B.R., Wells, L., Terns, R.M., and Terns, M.P. (2009). RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell 139, 945–956.
Hale, C.R., Cocozaki, A., Li, H., Terns, R.M., and Terns, M.P. (2014). Target RNA capture and cleavage by the Cmr type III-B CRISPR-Cas effector complex. Genes Dev 28, 2432–2443.
Han, W., Feng, X., and She, Q. (2017a). Reverse Gyrase Functions in Genome Integrity Maintenance by Protecting DNA Breaks In Vivo. IJMS 18, 1340.
Han, W., Li, Y., Deng, L., Feng, M., Peng, W., Hallstrøm, S., Zhang, J., Peng, N., Liang, Y.X., White, M.F., et al. (2017b). A type III-B CRISPR-Cas effector complex mediating massive target DNA destruction. Nucleic Acids Res 45, 1983–1993.
Han, W., Pan, S., López-Méndez, B., Montoya, G., and She, Q. (2017c). Allosteric regulation of Csx1, a type IIIB-associated CARF domain ribonuclease by RNAs carrying a tetraadenylate tail. Nucleic Acids Res 45, 10740–10750.
Han, W., Stella, S., Zhang, Y., Guo, T., Sulek, K., Peng-Lundgren, L., Montoya, G., and She, Q. (2018). A Type III-B Cmr effector complex catalyzes the synthesis of cyclic oligoadenylate second messengers by cooperative substrate binding. Nucleic Acids Res 46, 10319–10330.
He, F., Bhoobalan-Chitty, Y., Van, L.B., Kjeldsen, A.L., Dedola, M., Makarova, K.S., Koonin, E.V., Brodersen, D.E., and Peng, X. (2018). Anti-CRISPR proteins encoded by archaeal lytic viruses inhibit subtype I-D immunity. Nat Microbiol 3, 461–469.
Held, N.L., and Whitaker, R.J. (2009). Viral biogeography revealed by signatures in Sulfolobus islandicus genomes. Environ Microbiol 11, 457–466.
Hille, F., Richter, H., Wong, S.P., Bratovič, M., Ressel, S., and Charpentier, E. (2018). The biology of CRISPR-Cas: backward and forward. Cell 172, 1239–1259.
Hudaiberdiev, S., Shmakov, S., Wolf, Y.I., Terns, M.P., Makarova, K.S., and Koonin, E.V. (2017). Phylogenomics of Cas4 family nucleases. BMC Evol Biol 17, 232.
Ishino, Y., Shinagawa, H., Makino, K., Amemura, M., and Nakata, A. (1987). Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J Bacteriol 169, 5429–5433.
Jansen, R., Embden, J.D.A., Gaastra, W., and Schouls, L.M. (2002a). Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol 43, 1565–1575.
Jansen, R., van Embden, J.D.A., Gaastra, W., and Schouls, L.M. (2002b). Identification of a novel family of sequence repeats among prokaryotes. OMICS 6, 23–33.
Jaubert, C., Danioux, C., Oberto, J., Cortez, D., Bize, A., Krupovic, M., She, Q., Forterre, P., Prangishvili, D., and Sezonov, G. (2013). Genomics and genetics of Sulfolobus islandicus LAL14/1, a model hyperthermophilic archaeon. Open Biol 3, 130010.
Ji, X., Wang, D., and Gao, C. (2019). CRISPR editing-mediated antiviral immunity: a versatile source of resistance to combat plant virus infections. Sci China Life Sci 62, 1246–1249.
Jia, N., Jones, R., Sukenick, G., and Patel, D.J. (2019). Second messenger cA4 formation within the composite Csm1 palm pocket of type III-A CRISPR-Cas Csm complex and its release path. Mol Cell 75, 933–943. e6.
Jiang, W., Samai, P., and Marraffini, L.A. (2016). Degradation of phage transcripts by CRISPR-associated RNases enables type III CRISPR-Cas immunity. Cell 164, 710–721.
Jonuscheit, M., Martusewitsch, E., Stedman, K.M., and Schleper, C. (2003). A reporter gene system for the hyperthermophilic archaeon Sulfolobus solfataricus based on a selectable and integrative shuttle vector. Mol Microbiol 48, 1241–1252.
Kazlauskiene, M., Kostiuk, G., Venclovas, Č., Tamulaitis, G., and Siksnys, V. (2017). A cyclic oligonucleotide signaling pathway in type III CRISPR-Cas systems. Science 357, 605–609.
Kazlauskiene, M., Tamulaitis, G., Kostiuk, G., Venclovas, Č., and Siksnys, V. (2016). Spatiotemporal control of type III-A CRISPR-Cas immunity: coupling DNA degradation with the target RNA recognition. Mol Cell 62, 295–306.
Kim, Y.K., Kim, Y.G., and Oh, B.H. (2013). Crystal structure and nucleic acid-binding activity of the CRISPR-associated protein Csx1 of Pyrococcus furiosus. Proteins 81, 261–270.
Lau, C.H., Reeves, R., and Bolt, E.L. (2019). Adaptation processes that build CRISPR immunity: creative destruction, updated. Essays Biochem 63, 227–235.
Lee, H., Zhou, Y., Taylor, D.W., and Sashital, D.G. (2018). Cas4-dependent prespacer processing ensures high-fidelity programming of CRISPR arrays. Mol Cell 70, 48–59.e5.
León-Sobrino, C., Kot, W.P., and Garrett, R.A. (2016). Transcriptome changes in STSV2-infected Sulfolobus islandicus REY15A undergoing continuous CRISPR spacer acquisition. Mol Microbiol 99, 719–728.
Levy, A., Goren, M.G., Yosef, I., Auster, O., Manor, M., Amitai, G., Edgar, R., Qimron, U., and Sorek, R. (2015). CRISPR adaptation biases explain preference for acquisition of foreign DNA. Nature 520, 505–510.
Li, G., Liu, Y.G., and Chen, Y. (2019). Genome-editing technologies: the gap between application and policy. Sci China Life Sci 62, 1534–1538.
Li, M., Gong, L., Zhao, D., Zhou, J., and Xiang, H. (2017a). The spacer size of I-B CRISPR is modulated by the terminal sequence of the protospacer. Nucleic Acids Res 45, 4642–4654.
Li, M., Wang, R., and Xiang, H. (2014). Haloarcula hispanica CRISPR authenticates PAM of a target sequence to prime discriminative adaptation. Nucleic Acids Res 42, 7226–7235.
Li, Y., Pan, S., Zhang, Y., Ren, M., Feng, M., Peng, N., Chen, L., Liang, Y. X., and She, Q. (2016). Harnessing Type I and Type III CRISPR-Cas systems for genome editing. Nucleic Acids Res 44, e34.
Li, Y., Zhang, Y., Lin, J., Pan, S., Han, W., Peng, N., Liang, Y.X., and She, Q. (2017b). Cmr1 enables efficient RNA and DNA interference of a III-B CRISPR-Cas system by binding to target RNA and crRNA. Nucleic Acids Res 45, 11305–11314.
Lillestøl, R.K., Shah, S.A., Brügger, K., Redder, P., Phan, H., Christiansen, J., and Garrett, R.A. (2009). CRISPR families of the crenarchaeal genus Sulfolobus: bidirectional transcription and dynamic properties. Mol Microbiol 72, 259–272.
Lillestøl, R.K., Redder, P., Garrett, R.A., and Brügger, K. (2006). A putative viral defence mechanism in archaeal cells. Archaea 2, 59–72.
Lin, J., Feng, M., Zhang, H., and She, Q. (2020). Characterization of a novel type III CRISPR-Cas effector provides new insights into the allosteric activation and suppression of the Cas10 DNase. Cell Discov 6, 29.
Lintner, N.G., Kerou, M., Brumfield, S.K., Graham, S., Liu, H., Naismith, J.H., Sdano, M., Peng, N., She, Q., Copié, V., et al. (2011). Structural and functional characterization of an archaeal clustered regularly interspaced short palindromic repeat (CRISPR)-associated complex for antiviral defense (CASCADE). J Biol Chem 286, 21643–21656.
Liu, G., She, Q., and Garrett, R.A. (2016). Diverse CRISPR-Cas responses and dramatic cellular DNA changes and cell death in pKEF9-conjugated Sulfolobus species. Nucleic Acids Res 44, 4233–4242.
Liu, Q., Wang, C., Jiao, X., Zhang, H., Song, L., Li, Y., Gao, C., and Wang, K. (2019). Hi-TOM: a platform for high-throughput tracking of mutations induced by CRISPR/Cas systems. Sci China Life Sci 62, 1–7.
Liu, T., Li, Y., Wang, X., Ye, Q., Li, H., Liang, Y., She, Q., and Peng, N. (2015). Transcriptional regulator-mediated activation of adaptation genes triggers CRISPR de novo spacer acquisition. Nucleic Acids Res 43, 1044–1055.
Liu, T., Liu, Z., Ye, Q., Pan, S., Wang, X., Li, Y., Peng, W., Liang, Y., She, Q., and Peng, N. (2017). Coupling transcriptional activation of CRISPR-Cas system and DNA repair genes by Csa3a in Sulfolobus islandicus. Nucleic Acids Res 45, 8978–8992.
Majumdar, S., and Terns, M.P. (2018). CRISPR RNA-guided DNA cleavage by reconstituted Type I-A immune effector complexes. Extremophiles 23, 19–33.
Majumdar, S., Zhao, P., Pfister, N.T., Compton, M., Olson, S., Glover Iii, C. V.C., Wells, L., Graveley, B.R., Terns, R.M., and Terns, M.P. (2015). Three CRISPR-Cas immune effector complexes coexist in Pyrococcus furiosus. RNA 21, 1147–1158.
Makarova, K.S., Grishin, N.V., Shabalina, S.A., Wolf, Y.I., and Koonin, E. V. (2006). A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol Direct 1, 7.
Makarova, K.S., Anantharaman, V., Grishin, N.V., Koonin, E.V., and Aravind, L. (2014). CARF and WYL domains: ligand-binding regulators of prokaryotic defense systems. Front Genet 5, 102.
Makarova, K.S., Haft, D.H., Barrangou, R., Brouns, S.J.J., Charpentier, E., Horvath, P., Moineau, S., Mojica, F.J.M., Wolf, Y.I., Yakunin, A.F., et al. (2011). Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol 9, 467–477.
Makarova, K.S., Wolf, Y.I., Alkhnbashi, O.S., Costa, F., Shah, S.A., Saunders, S.J., Barrangou, R., Brouns, S.J.J., Charpentier, E., Haft, D. H., et al. (2015). An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol 13, 722–736.
Makarova, K.S., Wolf, Y.I., Iranzo, J., Shmakov, S.A., Alkhnbashi, O.S., Brouns, S.J.J., Charpentier, E., Cheng, D., Haft, D.H., Horvath, P., et al. (2020). Evolutionary classification of CRISPR-Cas systems: a burst of class 2 and derived variants. Nat Rev Microbiol 18, 67–83.
Mali, P., Yang, L., Esvelt, K.M., Aach, J., Guell, M., DiCarlo, J.E., Norville, J.E., and Church, G.M. (2013). RNA-guided human genome engineering via Cas9. Science 339, 823–826.
Manica, A., and Schleper, C. (2013). CRISPR-mediated defense mechanisms in the hyperthermophilic archaeal genus Sulfolobus. RNA Biol 10, 671–678.
Manica, A., Zebec, Z., Steinkellner, J., and Schleper, C. (2013). Unexpectedly broad target recognition of the CRISPR-mediated virus defence system in the archaeon Sulfolobus solfataricus. Nucleic Acids Res 41, 10509–10517.
Manica, A., Zebec, Z., Teichmann, D., and Schleper, C. (2011). In vivo activity of CRISPR-mediated virus defence in a hyperthermophilic archaeon. Mol Microbiol 80, 481–491.
Marraffini, L.A., and Sontheimer, E.J. (2008). CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 322, 1843–1845.
Marraffini, L.A., and Sontheimer, E.J. (2010). Self versus non-self discrimination during CRISPR RNA-directed immunity. Nature 463, 568–571.
McGinn, J., and Marraffini, L.A. (2019). Molecular mechanisms of CRISPR-Cas spacer acquisition. Nat Rev Microbiol 17, 7–12.
Medvedeva, S., Liu, Y., Koonin, E.V., Severinov, K., Prangishvili, D., and Krupovic, M. (2019). Virus-borne mini-CRISPR arrays are involved in interviral conflicts. Nat Commun 10, 5204.
Meng, X., Hu, X., Liu, Q., Song, X., Gao, C., Li, J., and Wang, K. (2018). Robust genome editing of CRISPR-Cas9 at NAG PAMs in rice. Sci China Life Sci 61, 122–125.
Mohanraju, P., Makarova, K.S., Zetsche, B., Zhang, F., Koonin, E.V., and van der Oost, J. (2016). Diverse evolutionary roots and mechanistic variations of the CRISPR-Cas systems. Science 353, aad5147.
Mojica, F.J.M., Díez-Villaseñor, C., García-Martínez, J., and Soria, E. (2005). Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol 60, 174–182.
Mojica, F.J.M., Juez, G., and Rodriguez-Valera, F. (1993). Transcription at different salinities of Haloferax mediterranei sequences adjacent to partially modified PstI sites. Mol Microbiol 9, 613–621.
Molina, R., Stella, S., Feng, M., Sofos, N., Jauniskis, V., Pozdnyakova, I., López-Méndez, B., She, Q., and Montoya, G. (2019). Structure of Csx1-cOA4 complex reveals the basis of RNA decay in Type III-B CRISPR-Cas. Nat Commun 10, 4302.
Mousaei, M., Deng, L., She, Q., and Garrett, R.A. (2016). Major and minor crRNA annealing sites facilitate low stringency DNA protospacer binding prior to Type I-A CRISPR-Cas interference in Sulfolobus. RNA Biol 13, 1166–1173.
Niewoehner, O., Garcia-Doval, C., Rostøl, J.T., Berk, C., Schwede, F., Bigler, L., Hall, J., Marraffini, L.A., and Jinek, M. (2017). Type III CRISPR-Cas systems produce cyclic oligoadenylate second messengers. Nature 548, 543–548.
Niewoehner, O., and Jinek, M. (2016). Structural basis for the endoribonuclease activity of the type III-A CRISPR-associated protein Csm6. RNA 22, 318–329.
Nuñez, J.K., Kranzusch, P.J., Noeske, J., Wright, A.V., Davies, C.W., and Doudna, J.A. (2014). Cas1-Cas2 complex formation mediates spacer acquisition during CRISPR-Cas adaptive immunity. Nat Struct Mol Biol 21, 528–534.
Osawa, T., Inanaga, H., Sato, C., and Numata, T. (2015). Crystal structure of the CRISPR-Cas RNA silencing Cmr complex bound to a target analog. Mol Cell 58, 418–430.
Pan, S., Li, Q., Deng, L., Jiang, S., Jin, X., Peng, N., Liang, Y., She, Q., and Li, Y. (2019). A seed motif for target RNA capture enables efficient immune defense by a type III-B CRISPR-Cas system. RNA Biol 16, 1166–1178.
Peng, W., Feng, M., Feng, X., Liang, Y.X., and She, Q. (2015). An archaeal CRISPR type III-B system exhibiting distinctive RNA targeting features and mediating dual RNA and DNA interference. Nucleic Acids Res 43, 406–417.
Peng, W., Li, H., Hallstrøm, S., Peng, N., Liang, Y.X., and She, Q. (2013). Genetic determinants of PAM-dependent DNA targeting and pre-crRNA processing in Sulfolobus islandicus. RNA Biol 10, 738–748.
Peng, X., Garrett, R.A., and She, Q.X. (2012). Archaeal viruses—novel, diverse and enigmatic. Sci China Life Sci 55, 422–433.
Plagens, A., Tripp, V., Daume, M., Sharma, K., Klingl, A., Hrle, A., Conti, E., Urlaub, H., and Randau, L. (2014). In vitro assembly and activity of an archaeal CRISPR-Cas type I-A Cascade interference complex. Nucleic Acids Res 42, 5125–5138.
Pourcel, C., Salvignol, G., and Vergnaud, G. (2005). CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology 151, 653–663.
Prangishvili, D., Bamford, D.H., Forterre, P., Iranzo, J., Koonin, E.V., and Krupovic, M. (2017). The enigmatic archaeal virosphere. Nat Rev Microbiol 15, 724–739.
Pyenson, N.C., and Marraffini, L.A. (2017). Type III CRISPR-Cas systems: when DNA cleavage just isn’t enough. Curr Opin Microbiol 37, 150–154.
Reeks, J., Sokolowski, R.D., Graham, S., Liu, H., Naismith, J.H., and White, M.F. (2013). Structure of a dimeric crenarchaeal Cas6 enzyme with an atypical active site for CRISPR RNA processing. Biochem J 452, 223–230.
Rollie, C., Graham, S., Rouillon, C., and White, M.F. (2018). Prespacer processing and specific integration in a Type I-A CRISPR system. Nucleic Acids Res 46, 1007–1020.
Rostøl, J.T., and Marraffini, L.A. (2019). Non-specific degradation of transcripts promotes plasmid clearance during type III-A CRISPR-Cas immunity. Nat Microbiol 4, 656–662.
Rouillon, C., Athukoralage, J.S., Graham, S., Grüschow, S., and White, M. F. (2018). Control of cyclic oligoadenylate synthesis in a type III CRISPR system. eLife 7, e36734.
Samai, P., Pyenson, N., Jiang, W., Goldberg, G.W., Hatoum-Aslan, A., and Marraffini, L.A. (2015). Co-transcriptional DNA and RNA cleavage during Type III CRISPR-Cas immunity. Cell 161, 1164–1174.
Schleper, C., Holz, I., Janekovic, D., Murphy, J., and Zillig, W. (1995). A multicopy plasmid of the extremely thermophilic archaeon Sulfolobus effects its transfer to recipients by mating. J Bacteriol 177, 4417–4426.
Semenova, E., Jore, M.M., Datsenko, K.A., Semenova, A., Westra, E.R., Wanner, B., van der Oost, J., Brouns, S.J.J., and Severinov, K. (2011). Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc Natl Acad Sci USA 108, 10098–10103.
Sensen, C.W., Klenk, H.P., Singh, R.K., Allard, G., Chan, C.C.Y., Liu, Q. Y., Penny, S.L., Young, F., Schenk, M.E., Gaasterland, T., et al. (1996). Organizational characteristics and information content of an archaeal genome: 156kb of sequence from Sulfolobus solfataricus P2. Mol Microbiol 22, 175–191.
Shah, S.A., Hansen, N.R., and Garrett, R.A. (2009). Distribution of CRISPR spacer matches in viruses and plasmids of crenarchaeal acidothermophiles and implications for their inhibitory mechanism. Biochem Soc Trans 37, 23–28.
Shao, Y., and Li, H. (2013). Recognition and cleavage of a nonstructured CRISPR RNA by its processing endoribonuclease Cas6. Structure 21, 385–393.
She, Q., Phan, H., Garrett, R.A., Albers, S.V., Stedman, K.M., and Zillig, W. (1998). Genetic profile of pNOB8 from Sulfolobus: the first conjugative plasmid from an archaeon. Extremophiles 2, 417–425.
She, Q., Singh, R.K., Confalonieri, F., Zivanovic, Y., Allard, G., Awayez, M.J., Chan-Weiher, C.C.Y., Groth Clausen, I., Curtis, B.A., De Moors, A., et al. (2001). The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc Natl Acad Sci USA 98, 7835–7840.
She, Q., Zhang, C., Deng, L., Peng, N., Chen, Z., and Liang, Y.X. (2009). Genetic analyses in the hyperthermophilic archaeon Sulfolobus islandicus. Biochem Soc Trans 37, 92–96.
Sheppard, N.F., Glover Iii, C.V.C., Terns, R.M., and Terns, M.P. (2016). The CRISPR-associated Csx1 protein of Pyrococcus furiosus is an adenosine-specific endoribonuclease. RNA 22, 216–224.
Shiimori, M., Garrett, S.C., Graveley, B.R., and Terns, M.P. (2018). Cas4 nucleases define the PAM, length, and orientation of DNA fragments integrated at CRISPR loci. Mol Cell 70, 814–824.e6.
Snyder, J.C., Bolduc, B., and Young, M.J. (2015). 40 Years of archaeal virology: expanding viral diversity. Virology 479–480, 369–378.
Sokolowski, R.D., Graham, S., and White, M.F. (2014). Cas6 specificity and CRISPR RNA loading in a complex CRISPR-Cas system. Nucleic Acids Res 42, 6532–6541.
Spilman, M., Cocozaki, A., Hale, C., Shao, Y., Ramia, N., Terns, R., Terns, M., Li, H., and Stagg, S. (2013). Structure of an RNA silencing complex of the CRISPR-Cas immune system. Mol Cell 52, 146–152.
Staals, R.H.J., Jackson, S.A., Biswas, A., Brouns, S.J.J., Brown, C.M., and Fineran, P.C. (2016). Interference-driven spacer acquisition is dominant over naive and primed adaptation in a native CRISPR-Cas system. Nat Commun 7, 12853.
Staals, R.H.J., Agari, Y., Maki-Yonekura, S., Zhu, Y., Taylor, D.W., van Duijn, E., Barendregt, A., Vlot, M., Koehorst, J.J., Sakamoto, K., et al. (2013). Structure and activity of the RNA-targeting Type III-B CRISPR-Cas complex of Thermus thermophilus. Mol Cell 52, 135–145.
Tamulaitis, G., Venclovas, Č., and Siksnys, V. (2017). Type III CRISPR-Cas immunity: major differences brushed aside. Trends Microbiol 25, 49–61.
Tang, T.H., Bachellerie, J.P., Rozhdestvensky, T., Bortolin, M.L., Huber, H., Drungowski, M., Elge, T., Brosius, J., and Hüttenhofer, A. (2002). Identification of 86 candidates for small non-messenger RNAs from the archaeon Archaeoglobus fulgidus. Proc Natl Acad Sci USA 99, 7536–7541.
Tang, T.H., Polacek, N., Zywicki, M., Huber, H., Brugger, K., Garrett, R., Bachellerie, J.P., and Hüttenhofer, A. (2005). Identification of novel non-coding RNAs as potential antisense regulators in the archaeon Sulfolobus solfataricus. Mol Microbiol 55, 469–481.
Taylor, D.W., Zhu, Y., Staals, R.H.J., Kornfeld, J.E., Shinkai, A., van der Oost, J., Nogales, E., and Doudna, J.A. (2015). Structures of the CRISPR-Cmr complex reveal mode of RNA target positioning. Science 348, 581–585.
Vestergaard, G., Garrett, R.A., and Shah, S.A. (2014). CRISPR adaptive immune systems of Archaea. RNA Biol 11, 156–167.
Wang, H., Peng, N., Shah, S.A., Huang, L., and She, Q. (2015). Archaeal extrachromosomal genetic elements. Microbiol Mol Biol Rev 79, 117–152.
Xue, C., Zhang, H., Lin, Q., Fan, R., and Gao, C. (2018). Manipulating mRNA splicing by base editing in plants. Sci China Life Sci 61, 1293–1300.
Yosef, I., Goren, M.G., and Qimron, U. (2012). Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res 40, 5569–5576.
You, L., Ma, J., Wang, J., Artamonova, D., Wang, M., Liu, L., Xiang, H., Severinov, K., Zhang, X., and Wang, Y. (2019). Structure studies of the CRISPR-Csm complex reveal mechanism of co-transcriptional interference. Cell 176, 239–253.e16.
Zebec, Z., Manica, A., Zhang, J., White, M.F., and Schleper, C. (2014). CRISPR-mediated targeted mRNA degradation in the archaeon Sulfolobus solfataricus. Nucleic Acids Res 42, 5280–5288.
Zhang, C., Guo, L., Deng, L., Wu, Y., Liang, Y., Huang, L., and She, Q. (2010). Revealing the essentiality of multiple archaeal pcna genes using a mutant propagation assay based on an improved knockout method. Microbiology 156, 3386–3397.
Zhang, J., Graham, S., Tello, A., Liu, H., and White, M.F. (2016). Multiple nucleic acid cleavage modes in divergent type III CRISPR systems. Nucleic Acids Res 44, 1789–1799.
Zhang, J., Kasciukovic, T., and White, M.F. (2012a). The CRISPR associated protein Cas4 is a 5′ to 3′ DNA exonuclease with an iron-sulfur cluster. PLoS ONE 7, e47232.
Zhang, J., Rouillon, C., Kerou, M., Reeks, J., Brugger, K., Graham, S., Reimann, J., Cannone, G., Liu, H., Albers, S.V., et al. (2012b). Structure and mechanism of the CMR complex for CRISPR-mediated antiviral immunity. Mol Cell 45, 303–313.
Zhang, Y., Lin, J., Feng, M., and She, Q. (2018). Molecular mechanisms of III-B CRISPR-Cas systems in archaea. Emerg Top Life Sci 2, 483–491.
Zhang, Z., Pan, S., Liu, T., Li, Y., and Peng, N. (2019). Cas4 Nucleases Can Effect Specific Integration of CRISPR Spacers. J Bacteriol 201, pii: e00747–00718.
Zhu, Y., Klompe, S.E., Vlot, M., van der Oost, J., and Staals, R.H.J. (2018). Shooting the messenger: RNA-targetting CRISPR-Cas systems. Biosci Rep 38.
Zink, I.A., Pfeifer, K., Wimmer, E., Sleytr, U.B., Schuster, B., and Schleper, C. (2019). CRISPR-mediated gene silencing reveals involvement of the archaeal S-layer in cell division and virus infection. Nat Commun 10, 4797.
Acknowledgements
This work was supported by grants from the Chinese National Transgenic Science and Technology Program (2019ZX08010003 to QS), the National Natural Science Foundation of China (31771380 to QS), the Qingdao Applied Research Fund for postdoctoral researchers (62450079311107 to ZY), and the State Key Laboratory of Microbial Technology and Shandong University.
Author information
Authors and Affiliations
Corresponding author
Additional information
Compliance and ethics
The author(s) declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Yu, Z., Jiang, S., Wang, Y. et al. CRISPR-Cas adaptive immune systems in Sulfolobales: genetic studies and molecular mechanisms. Sci. China Life Sci. 64, 678–696 (2021). https://doi.org/10.1007/s11427-020-1745-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11427-020-1745-0