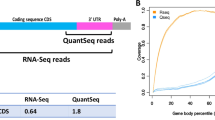
Abstract
RNA-Seq promises to be used in clinical settings as a gene-expression profiling tool; however, questions about its variability and biases remain and need to be addressed. Thus, RNA controls with known concentrations and sequence identities originally developed by the External RNA Control Consortium (ERCC) for microarray and qPCR platforms have recently been proposed for RNA-Seq platforms, but only with a limited number of samples. In this study, we report our analysis of RNA-Seq data from 92 ERCC controls spiked in a diverse collection of 447 RNA samples from eight ongoing studies involving five species (human, rat, mouse, chicken, and Schistosoma japonicum) and two mRNA enrichment protocols, i.e., poly(A) and RiboZero. The entire collection of datasets consisted of 15650143175 short sequence reads, 131603796 (i.e., 0.84%) of which were mapped to the 92 ERCC references. The overall ERCC mapping ratio of 0.84% is close to the expected value of 1.0% when assuming a 2.0% mRNA fraction in total RNA, but showed a difference of 2.8-fold across studies and 4.3-fold among samples from the same study with one tissue type. This level of fluctuation may prevent the ERCC controls from being used for cross-sample normalization in RNA-Seq. Furthermore, we observed striking biases of quantification between poly(A) and RiboZero which are transcript-specific. For example, ERCC-00116 showed a 7.3-fold under-enrichment in poly(A) compared to RiboZero. Extra care is needed in integrative analysis of multiple datasets and technical artifacts of protocol differences should not be taken as true biological findings.
Article PDF
Similar content being viewed by others
Avoid common mistakes on your manuscript.
References
Xuan J, Yu Y, Qing T, et al. Next-generation sequencing in the clinic: promises and challenges. Cancer Lett, 2012, doi: 10.1016/j.canlet.2012.11.025
Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet, 2009, 10: 57–63
Mutz K, Heilkenbrinker A, Lönne M, et al. Transcriptome analysis using next-generation sequencing. Curr Opin Biotechnol, 2012, 24: 1–9
Nagalakshmi U, Wang Z, Waern K, et al. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science, 2008, 320: 1344–1349
Cloonan N, Forrest AR, Kolle G, et al. Stem cell transcriptome profiling via massive-scale mRNA sequencing. Nat Methods, 2008, 5: 613–619
Marioni J C, Mason C E, Mane S M, et al. RNA-Seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res, 2008, 18: 1509–1517
McIntyre L M, Lopiano K K, Morse A M, et al. RNA-Seq: technical variability and sampling. BMC Genomics, 2011, 12: 293
Schwartz S, Oren R, Ast G. Detection and removal of biases in the analysis of next-generation sequencing reads. PLoS ONE, 2011, 6: e16685
Zheng W, Chung L M, Zhao H. Bias detection and correction in RNA-sequencing data. BMC Bioinformatics, 2011, 12: 290
Zhang J X, Coombes K R. Sources of variation in false discovery rate estimation include sample size, correlation, and inherent differences between groups. BMC Bioinformatics, 2012, 13: S1
Tong W, Lucas AB, Shippy R, et al. Evaluation of external RNA controls for the assessment of microarray performance. Nat Biotechnol, 2006, 24: 1132–1139
Kralj J G, Salit M L. Characterization of in vitro transcription amplification linearity and variability in the low copy number regime using External RNA Control Consortium (ERCC) Spike-ins. Anal Bioanal Chem, 2013, 405: 315–320
Baker S C, Bauer S R, Beyer R P, et al. The External RNA Controls Consortium: a progress report. Nat Methods, 2005, 2: 731–734
Devonshire A S, Elaswarapu R, Foy C A. Evaluation of external RNA controls for the standardisation of gene expression biomarker measurements. BMC Genomics, 2010, 11: 662
Jiang L, Schlesinger F, Davis C A, et al. Synthetic spike-in standards for RNA-Seq experiments. Genome Res, 2011, 21: 1543–1551
Loven J, Orlando D A, Sigova A A, et al. Revisiting global gene expression analysis. Cell, 2012, 151: 476–482
Zook J M, Samarov D, McDaniel J, et al. Synthetic spike-in standards improve run-specific systematic error analysis for DNA and RNA sequencing. PLoS ONE, 2012, 7: e41356
Warrington J A, Corbisier P, Feilotter H, et al. Use of external RNA controls in gene expression assays: approved guideline. CLSI document MM16-A (ISBN 1-56238-617-4), Wayne, Peennsylvania, USA, 2006
Langmead B, Salzberg S L. Fast gapped-read alignment with bowtie 2. Nat Methods, 2012, 9: 357–359
Ramirez-Gonzalez R H, Bonnal R, Caccamo M, et al. Bio-samtools: ruby bindings for samtools, a library for accessing bam files containing high-throughput sequence alignments. Source Code Biol Med, 2012, 7: 6
Quinlan A R, Hall I M. Bedtools: a flexible suite of utilities for comparing genomic features. Bioinformatics, 2010, 26: 841–842
Shi L, Reid L H, Jones W D, et al. The MicroArray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat Biotechnol, 2006, 24: 1151–1161
Shi L, Campbell G, Jones W D, et al. The MicroArray Quality Control (MAQC)-II study of common practices for the development and validation of microarray-based predictive models. Nat Biotechnol, 2010, 28: 827–838
Author information
Authors and Affiliations
Corresponding author
Additional information
Contributed equally to this work
This article is published with open access at Springerlink.com
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 2.0 International License (https://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Qing, T., Yu, Y., Du, T. et al. mRNA enrichment protocols determine the quantification characteristics of external RNA spike-in controls in RNA-Seq studies. Sci. China Life Sci. 56, 134–142 (2013). https://doi.org/10.1007/s11427-013-4437-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11427-013-4437-9