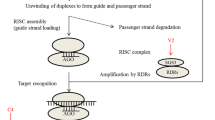
Abstract
Antiviral defense is one of the important roles of RNA silencing in plants. Virus-derived small interfering RNAs (vsiRNAs) are found in the infected host cells, indicating that the host RNA silencing machinery can target viral RNAs for destruction. With the development of high-throughput sequencing of vsiRNAs, recent genetic studies have shed light on the origin and composition of vsiRNAs and their potential functions in the regulation of gene expression. Here, we briefly describe the origin and biogenesis of vsiRNAs, and review the recent discoveries regarding vsiRNA-mediated RNA silencing of viral genomes and host transcripts. This will better our understanding of virus pathogenicity and RNA silencing-related host-pathogen interactions in plants.
Article PDF
Similar content being viewed by others
Avoid common mistakes on your manuscript.
References
Chen X. Small RNAs—secrets and surprises of the genome. Plant J, 2010, 61: 941–958
Ghildiyal M, Zamore P D. Small silencing RNAs: an expanding universe. Nat Rev Genet, 2009, 10: 94–108
Chapman E J, Carrington J C. Specialization and evolution of endogenous small RNA pathways. Nat Rev Genet, 2007, 8: 884–896
Hamilton A J, Baulcombe D C. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science, 1999, 286: 950–952
Burgyan J. Role of silencing suppressor proteins. Methods Mol Biol, 2008, 451: 69–79
Ding S W, Voinnet O. Antiviral immunity directed by small RNAs. Cell, 2007, 130: 413–426
Voinnet O, Pinto Y M, Baulcombe D C. Suppression of gene silencing: a general strategy used by diverse DNA and RNA viruses of plants. Proc Natl Acad Sci USA, 1999, 96: 14147–14152
Szittya G, Moxon S, Pantaleo V, et al. Structural and functional analysis of viral siRNAs. PLoS Pathog, 2010, 6: e1000838
Qu F. Antiviral role of plant-encoded RNA-dependent RNA polymerases revisited with deep sequencing of small interfering RNAs of virus origin. Mol Plant Microbe Interact, 2010, 23: 1248–1252
Donaire L, Wang Y, Gonzalez-Ibeas D, et al. Deep-sequencing of plant viral small RNAs reveals effective and widespread targeting of viral genomes. Virology, 2009, 392: 203–214
Donaire L, Barajas D, Martinez-Garcia B, et al. Structural and genetic requirements for the biogenesis of tobacco rattle virus-derived small interfering RNAs. J Virol, 2008, 82: 5167–5177
Pantaleo V, Szittya G, Burgyan J. Molecular Bases of Viral RNA Targeting by Viral Small Interfering RNA-Programmed RISC. J Virol, 2007, 81: 3797–3806
Molnar A, Csorba T, Lakatos L, et al. Plant Virus-derived small interfering RNAs originate predominantly from highly structured single-stranded viral RNAs. J Virol, 2005, 79: 7812–7818
Wang X B, Wu Q, Ito T, et al. RNAi-mediated viral immunity requires amplification of virus-derived siRNAs in Arabidopsis thaliana. Proc Natl Acad Sci USA, 2010, 107: 484–489
Garcia-Ruiz H, Takeda A, Chapman E J, et al. Arabidopsis RNA-dependent RNA polymerases and Dicer-like proteins in antiviral defense and small interfering RNA biogenesis during Turnip Mosaic Virus infection. Plant Cell, 2010, 22: 481–496
Qi X, Bao F S, Xie Z. Small RNA deep sequencing reveals role for Arabidopsis thaliana RNA-dependent RNA polymerases in viral siRNA biogenesis. PLoS ONE, 2009, 4: e4971
Azevedo J, Garcia D, Pontier D, et al. Argonaute quenching and global changes in Dicer homeostasis caused by a pathogen-encoded GW repeat protein. Genes Dev, 2010, 24: 904–915
Zhang X, Yuan Y R, Pei Y, et al. Cucumber mosaic virus-encoded 2b suppressor inhibits Arabidopsis Argonaute1 cleavage activity to counter plant defense. Genes Dev, 2006, 20: 3255–3268
Pantaleo V, Burgyan J. Cymbidium ringspot virus harnesses RNA silencing to control the accumulation of virus parasite satellite RNA. J Virol, 2008, 82: 11851–11858
Golem S, Culver J N. Tobacco mosaic virus induced alterations in the gene expression profile of Arabidopsis thaliana. Mol Plant Microbe Interact, 2003, 16: 681–688
Whitham S A, Yang C, Goodin M M. Global impact: elucidating plant responses to viral infection. Mol Plant Microbe Interact, 2006, 19: 1207–1215
Agudelo-Romero P, Carbonell P, de la Iglesia F, et al. Changes in the gene expression profile of Arabidopsis thaliana after infection with Tobacco etch virus. Virol J, 2008, 5: 92
Havelda Z, Varallyay E, Valoczi A, et al. Plant virus infection-induced persistent host gene downregulation in systemically infected leaves. Plant J, 2008, 55: 278–288
Ahlquist P. RNA-dependent RNA polymerases, viruses, and RNA silencing. Science, 2002, 296: 1270–1273
den Boon J A, Ahlquist P. Organelle-like membrane compartmentalization of positive-strand RNA virus replication factories. Annu Rev Microbiol, 2010, 64: 241–256
Moissiard G, Voinnet O. RNA silencing of host transcripts by cauliflower mosaic virus requires coordinated action of the four Arabidopsis Dicer-like proteins. Proc Natl Acad Sci USA, 2006, 103: 19593–19598
Vanitharani R, Chellappan P, Fauquet C M. Geminiviruses and RNA silencing. Trends Plant Sci, 2005, 10: 144–151
Chellappan P, Vanitharani R, Fauquet C M. Short Interfering RNA accumulation correlates with host recovery in DNA virus-Infected hosts, and gene silencing targets specific viral sequences. J Virol, 2004, 78: 7465–7477
Fusaro A F, Matthew L, Smith N A, et al. RNA interference-inducing hairpin RNAs in plants act through the viral defence pathway. EMBO Rep, 2006, 7: 1168–1175
Bouche N, Lauressergues D, Gasciolli V, et al. An antagonistic function for Arabidopsis DCL2 in development and a new function for DCL4 in generating viral siRNAs. EMBO J, 2006, 25: 3347–3356
Deleris A, Gallego-Bartolome J, Bao J, et al. Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science, 2006, 313: 68–71
Blevins T, Rajeswaran R, Shivaprasad P V, et al. Four plant Dicers mediate viral small RNA biogenesis and DNA virus induced silencing. Nucleic Acids Res, 2006, 34: 6233–6246
Yoshikawa M, Peragine A, Park M Y, et al. A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev, 2005, 19: 2164–2175
Vazquez F, Vaucheret H, Rajagopalan R, et al. Endogenous transacting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol Cell, 2004, 16: 69–79
Peragine A, Yoshikawa M, Wu G, et al. SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of transacting siRNAs in Arabidopsis. Genes Dev, 2004, 18: 2368–2379
Baulcombe D. RNA silencing in plants. Nature, 2004, 431: 356–363
Harvey J J W, Lewsey M G, Patel K, et al. An antiviral defense role of AGO2 in plants. PLoS ONE, 2011, 6: e14639
Takeda A, Iwasaki S, Watanabe T, et al. The mechanism selecting the guide strand from small RNA duplexes is different among Argonaute proteins. Plant Cell Physiol, 2008, 49: 493–500
Qu F, Ye X, Morris T J. Arabidopsis DRB4, AGO1, AGO7, and RDR6 participate in a DCL4-initiated antiviral RNA silencing pathway negatively regulated by DCL1. Proc Natl Acad Sci USA, 2008, 105: 14732–14737
Wang X B, Jovel J, Udomporn P, et al. The 21-nucleotide, but not 22-nucleotide, viral secondary small interfering RNAs direct potent antiviral defense by two cooperative Argonautes in Arabidopsis thaliana. Plant Cell, 2011, 23: 1625–1638
Montgomery T A, Howell M D, Cuperus J T, et al. Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell, 2008, 133: 128–141
Mi S, Cai T, Hu Y, et al. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell, 2008, 133: 116–127
Simon-Mateo C, Garcia J A. MicroRNA-guided processing impairs Plum pox virus replication, but the virus readily evolves to escape this silencing mechanism. J Virol, 2006, 80: 2429–2436
Li H, Li W X, Ding S W. Induction and suppression of RNA silencing by an animal virus. Science, 2002, 296: 1319–1321
Diaz-Pendon J A, Li F, Li W X, et al. Suppression of antiviral silencing by cucumber mosaic virus 2b protein in Arabidopsis is associated with drastically reduced accumulation of three classes of viral small interfering RNAs. Plant Cell 2007, 19: 2053–2063
Martin-Hernandez A M, Baulcombe D C. Tobacco rattle virus 16-kilodalton protein encodes a suppressor of RNA silencing that allows transient viral entry in meristems. J Virol, 2008, 82: 4064–4071
Du Q S, Duan C G, Zhang Z H, et al. DCL4 targets Cucumber mosaic virus satellite RNA at novel secondary structures. J Virol, 2007, 81: 9142–9151
Zhu H, Duan C G, Hou W N, et al. Satellite RNA-derived satsiR-12 targeting the 3’UTR of Cucumber mosaic virus triggers viral RNAs for degradation. J Virol, 2011, 85: 13384–13397
Hou W N, Duan C G, Fang R X, et al. Satellite RNA reduces expression of the 2b suppressor protein resulting in the attenuation of symptoms caused by Cucumber mosaic virus infection. Mol Plant Pathol, 2011, 12: 595–605
Alvarado V, Scholthof H B. Plant responses against invasive nucleic acids: RNA silencing and its suppression by plant viral pathogens. Semin Cell Dev Biol, 2009, 20: 1032–1040
Díaz-Pendón J A, Ding S W. Direct and indirect roles of viral suppressors of RNA silencing in pathogenesis. Annu Rev Phytopathol, 2008, 46: 303–326
Dunoyer P, Voinnet O. The complex interplay between plant viruses and host RNA-silencing pathways. Curr Opin Plant Biol, 2005, 8: 415–423
Wang M B. On the role of RNA silencing in the pathogenicity and evolution of viroids and viral satellites. Proc Natl Acad Sci USA, 2004, 101: 3275–3280
Devic M, Jaegle M, Baulcombe D. Symptom production on tobacco and tomato is determined by two distinct domains of the satellite RNA of cucumber mosaic virus (strain Y). J Gen Virol, 1989, 70: 2765–2774
Masuta C, Takanami Y. Determination of sequence and structural requirements for pathogenicity of a cucumber mosaic virus satellite RNA (Y-satRNA). Plant Cell, 1989, 1: 1165–1173
Jaegle M, Devic M, Longstaff M, et al. Cucumber mosaic virus satellite RNA (Y strain): analysis of sequences which affect yellow mosaic symptoms on tobacco. J Gen Virol, 1990, 71: 1905–1912
Kuwata S, Masuta C, Takanami Y. Reciprocal phenotype alterations between two satellite RNAs of cucumber mosaic virus. J Gen Virol, 1991, 72: 2385–2389
Smith N A, Eamens A L, Wang M B. Viral small interfering RNAs target host genes to mediate disease symptoms in plants. PLoS Pathog, 2011, 7: e1002022
Li W X, Ding S W. Viral suppressors of RNA silencing. Curr Opin Biotechnol, 2001, 12: 150–154
Moissiard G, Voinnet O. Viral suppression of RNA silencing in plants. Mol Plant Pathol, 2004, 5: 71–82
Roth B M, Pruss G J, Vance V B. Plant viral suppressors of RNA silencing. Virus Res, 2004, 102: 97–108
Shimura H, Pantaleo V, Ishihara T, et al. A viral satellite RNA induces yellow symptoms on tobacco by targeting a gene involved in chlorophyll biosynthesis using the RNA silencing machinery. PLoS Pathog, 2011, 7: e1002021
Manfre A. Importance of coat protein and RNA silencing in satellite RNA/virus interactions. Virology, 2008, 379: 161–167
Zhang F, Simon A E. Enhanced viral pathogenesis associated with a virulent mutant virus or a virulent satellite RNA correlates with reduced virion accumulation and abundance of free coat protein. Virology, 2003, 312: 8–13
Raja P, Sanville B C, Buchmann R C, et al. Viral genome methylation as an epigenetic defense against geminiviruses. J Virol, 2008, 82: 8997–9007
Yang X, Xie Y, Raja P, et al. Suppression of methylation-mediated transcriptional gene silencing by βC1-SAHH protein interaction during geminivirus-betasatellite infection. PLoS Pathog, 2011, 7: e1002329
Blevins T, Rajeswaran R, Aregger M, et al. Massive production of small RNAs from a non-coding region of Cauliflower mosaic virus in plant defense and viral counter-defense. Nucleic Acids Res, 2011, 39: 5003–5014
Author information
Authors and Affiliations
Corresponding author
Additional information
This article is published with open access at Springerlink.com
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 2.0 International License (https://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Zhu, H., Guo, H. The role of virus-derived small interfering RNAs in RNA silencing in plants. Sci. China Life Sci. 55, 119–125 (2012). https://doi.org/10.1007/s11427-012-4281-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11427-012-4281-3