Abstract
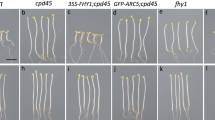
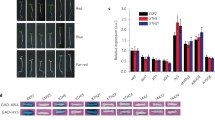
In this study, we show that CIPK14, a stress responsive CBL-interacting protein kinase gene, is involved in phytochrome A-mediated far-red light inhibition of greening in Arabidopsis seedlings. The CIPK14-impairment mutant cipk14 grown in continuous far-red (FR) light did not show greening when exposed to white light illumination for 15 h. By contrast, the FR-grown phytochrome A null mutant phyA greened within 0.5 h of exposure to white light. Although greening of Col-4 (wild-type) was not completely abolished by FR, it exhibited a significantly decreased greening capacity compared with that of phyA. Further analyses demonstrated that the expression of protochlorophyllide reductase (POR) genes was correlated with the greening ability of the genotypes. In addition, CIPK14 appeared to be regulated by both the circadian clock and PhyA. Taken together, these results suggest that CIPK14 plays a role in PhyA-mediated FR inhibition of seedling greening, and that a Ca-related kinase may be involved in a previously undefined branch point in the phytochrome A signaling pathway.
Similar content being viewed by others
References
Trewavas A J, Knight M R. Mechanical signalling, calcium and plant form. Plant Mol Biol, 1994, 26: 1329–1341 1:CAS:528:DyaK2MXjvFelu7g%3D, 10.1007/BF00016478, 7858194
Weinl S, Kudla J. The CBL-CIPK Ca2+-decoding signalling network: function and perspectives. New Phytol, 2009, 184: 517–528 1:CAS:528:DC%2BD1MXhsVOgs7vE, 10.1111/j.1469-8137.2009.02938.x, 19860013
McAinsh M R, Pittman J K. Shaping the calcium signature. New Phytol, 2009, 181: 275–294 1:CAS:528:DC%2BD1MXhvV2ns7k%3D, 10.1111/j.1469-8137.2008.02682.x, 19121028
Chung E, Park J M, Oh S K, et al. Molecular and biochemical characterization of the Capsicum annuum calcium-dependent protein kinase 3 (CaCDPK3) gene induced by abiotic and biotic stresses. Planta, 2004, 220: 286–295 1:CAS:528:DC%2BD2cXhtVensrfI, 10.1007/s00425-004-1372-9, 15449060
Ludwig A A, Saitoh H, Felix G, et al. Ethylene-mediated cross-talk between calcium-dependent protein kinase and MAPK signaling controls stress responses in plants. Proc Natl Acad Sci USA, 2005, 102: 10736–10741 1:CAS:528:DC%2BD2MXntVShsrs%3D, 10.1073/pnas.0502954102, 16027369
Frohnmeyer H, Loyall L, Blatt MR, et al. Millisecond UV-B irradiation evokes prolonged elevation of cytosolic-free Ca2+ and stimulates gene expression in transgenic parsley cell cultures. Plant J, 1999, 20: 109–117 1:CAS:528:DyaK1MXnsVGqsr8%3D, 10.1046/j.1365-313X.1999.00584.x, 10571870
Kim K N, Cheong Y H, Grant J J, et al. CIPK3, a calcium sensor-associated protein kinase that regulates abscisic acid and cold signal transduction in Arabidopsis. Plant Cell, 2003, 15: 411–423 1:CAS:528:DC%2BD3sXhtlOmtbo%3D, 10.1105/tpc.006858, 12566581
Trewavas A J, Malhó R. Ca2+ signalling in plant cells: the big network! Curr Opin Plant Biol, 1998, 1: 428–433 1:CAS:528:DyaK1cXnt1Oisrc%3D, 10.1016/S1369-5266(98)80268-9, 10066614
Nie X, Durnin D C, Igamberdiev A U, et al. Cytosolic calcium is involved in the regulation of barley hemoglobin gene expression. Planta, 2006, 223: 542–549 1:CAS:528:DC%2BD28XivFWjsbs%3D, 10.1007/s00425-005-0094-y, 16177910
Kolukisaoglu U, Weinl S, Blazevic D, et al. Calcium sensors and their interacting protein kinases: genomics of the Arabidopsis and rice CBL-CIPK signaling networks. Plant Physiol, 2004, 134: 43–58 1:CAS:528:DC%2BD2cXhtVagtbo%3D, 10.1104/pp.103.033068, 14730064
Ishitani M, Liu J, Halfter U, et al. SOS3 function in plant salt tolerance requires N-myristoylation and calcium binding. Plant Cell, 2000, 12: 1667–1678 1:CAS:528:DC%2BD3cXnsVyltrk%3D, 10.1105/tpc.12.9.1667, 11006339
Nagae M, Nozawa A, Koizumi N, et al. The crystal structure of the novel calcium-binding protein AtCBL2 from Arabidopsis thaliana. J Biol Chem, 2003, 278: 42240–42246 1:CAS:528:DC%2BD3sXot1amtbc%3D, 10.1074/jbc.M303630200, 12871972
Halfter U, Ishitani M, Zhu J K. The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proc Natl Acad Sci USA, 2000, 97: 3735–3740 1:CAS:528:DC%2BD3cXitlajsrg%3D, 10.1073/pnas.040577697, 10725350
Hrabak E M, Chan C W, Gribskov M, et al. The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol, 2003, 132: 666–680 1:CAS:528:DC%2BD3sXkslertr8%3D, 10.1104/pp.102.011999, 12805596
Zhu J K. Salt and drought stress signal transduction in plants. Annu Rev Plant Biol, 2002, 53: 247–273 1:CAS:528:DC%2BD38XlsVWhtbc%3D, 10.1146/annurev.arplant.53.091401.143329, 12221975
Pandey G K, Grant J J, Cheong Y H, et al. Calcineurin-B-like protein CBL9 interacts with target kinase CIPK3 in the regulation of ABA response in seed germination. Molecular Plant, 2008, 1: 238–248 1:CAS:528:DC%2BD1cXoslyks7w%3D, 10.1093/mp/ssn003, 19825536
Qin Y Z, Li X, Guo M, et al. Regulation of salt and ABA responses by CIPK14, a calcium sensor interacting protein kinase in Arabidopsis. Sci China Ser C-Life Sci, 2008, 51: 391–401 1:CAS:528:DC%2BD1cXoslSksLg%3D, 10.1007/s11427-008-0059-z
Johnson C H, Knight M R, Kondo T, et al. Circadian oscillations of cytosolic and chloroplastic free calcium in plants. Science, 1995, 269: 1863–1865 1:CAS:528:DyaK2MXotlKqtLg%3D, 10.1126/science.7569925, 7569925
Webb A A R. The physiology of circadian rhythms in plants. New Phytol, 2003, 160: 281–303 1:CAS:528:DC%2BD3sXpt1Ogsbo%3D, 10.1046/j.1469-8137.2003.00895.x
Dodd A N, Love J, Webb A A R. The plant clock shows its metal: circadian regulation of cytosolic free Ca2+. Trends Plant Sci, 2005, 10: 15–21 1:CAS:528:DC%2BD2MXitFarug%3D%3D, 10.1016/j.tplants.2004.12.001, 15642519
Eckardt N A. Circadian regulation of Ca2+ signalling. Plant Cell, 2007, 19: 3317
Love J, Dodd A N, Webb A A R. Circadian and diurnal calcium oscillations encode photoperiodic information in Arabidopsis. Plant Cell, 2004, 16: 956–966 1:CAS:528:DC%2BD2cXjt1ynsb8%3D, 10.1105/tpc.020214, 15031410
Harmer S L, Hogenesch J B, Straume M, et al. Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science, 2000, 290: 2110–2113 1:CAS:528:DC%2BD3cXptVyisrw%3D, 10.1126/science.290.5499.2110, 11118138
Edwards K D, Anderson P E, Hall A, et al. FLOWERING LOCUS C mediates natural variation in the high-temperature response of the Arabidopsis circadian clock. Plant Cell, 2006, 18: 639–650 1:CAS:528:DC%2BD28XisFahur4%3D, 10.1105/tpc.105.038315, 16473970
Covington M F, Harmer S L. The circadian clock regulates auxin signaling and responses in Arabidopsis. PLoS Biol, 2007, 5: e222 10.1371/journal.pbio.0050222, 17683202
Michael T P, Mockler T C, Breton G, et al. Network discovery pipeline elucidates conserved time-of-day-specific cis-regulatory modules. PLoS Genet, 2008, 4: e14 10.1371/journal.pgen.0040014, 18248097
Mizuno T, Yamashino T. Comparative transcriptome of diurnally oscillating genes and hormone-responsive genes in Arabidopsis thaliana: insight into circadian clock-controlled daily responses to common ambient stresses in plants. Plant Cell Physiol, 2008, 49: 481–487 1:CAS:528:DC%2BD1cXkvVWjsro%3D, 10.1093/pcp/pcn008, 18202002
Xu X, Hotta C T, Dodd A N, et al. Distinct light and clock modulation of cytosolic free Ca2+ oscillations and rhythmic CHLOROPHYLL A/B BINDING PROTEIN2 promoter activity in Arabidopsis. Plant Cell, 2007, 19: 3474–3490 1:CAS:528:DC%2BD1cXns1emsw%3D%3D, 10.1105/tpc.106.046011, 17982000
Schafer E, Nagy F, eds. Photomorphogenesis in Plants and Bacteria. 3rd ed. Dordrecht: Springer, 2006
Casson S A, Franklin K A, Gray J E, et al. Phytochrome B and PIF4 regulate stomatal development in response to light quantity. Curr Biol. 2009, 19: 229–234 1:CAS:528:DC%2BD1MXhs1aisL0%3D, 10.1016/j.cub.2008.12.046, 19185498
Kang C Y, Lian H L, Wang F F, et al. Cryptochromes, phytochromes, and COP1 regulate light controlled stomatal development in Arabidopsis. Plant Cell, 2009, 21: 2624–2641 1:CAS:528:DC%2BD1MXhsVejtbfM, 10.1105/tpc.109.069765, 19794114
Somers D E, Devlin P F, Kay S A. Phytochromes and cryptochromes in the entrainment of the Arabidopsis circadian clock. Science, 1998, 282: 1488–1494 1:CAS:528:DyaK1cXns1yqtL4%3D, 10.1126/science.282.5393.1488, 9822379
Sharrock R A, Quail P H. Novel phytochrome sequences in Arabidopsis thaliana: structure, evolution, and differential expression of a plant regulatory photoreceptor family. Genes Dev, 1989, 3: 1745–1757 1:CAS:528:DyaK3cXktFyltro%3D, 10.1101/gad.3.11.1745, 2606345
Clack T, Mathews S, Sharrock R A. The phytochrome apoprotein family in Arabidopsis is encoded by five genes: the sequence and expression of PHYD and PHYE. Plant Mol Biol, 1994, 25: 413–427 1:CAS:528:DyaK2cXmt1egtLs%3D, 10.1007/BF00043870, 8049367
Franklin K A, Quail P H. Phytochrome functions in Arabidopsis development. J Exp Bot, 2010, 61: 11–24 1:CAS:528:DC%2BD1MXhsFGnsr7J, 10.1093/jxb/erp304, 19815685
Wang H, Deng X W. Dissecting the phytochrome A-dependent signaling network in higher plants. Trends Plant Sci, 2003, 8: 172–178 1:CAS:528:DC%2BD3sXjtFeqs7k%3D, 10.1016/S1360-1385(03)00049-9, 12711229
Clough R C, Vierstra R D. Phytochrome degradation. Plant Cell Environ, 1997, 20: 713–721 1:CAS:528:DyaK2sXmt1Sqs7Y%3D, 10.1046/j.1365-3040.1997.d01-107.x
Sharrock R A, Clack T. Patterns of expression and normalized levels of the five Arabidopsis phytochromes. Plant Physiol, 2002, 130: 442–456 1:CAS:528:DC%2BD38XntFOrtL0%3D, 10.1104/pp.005389, 12226523
Guo H, Mockler T, Duong H, et al. SUB1, an Arabidopsis Ca2+-binding protein involved in cryptochrome and phytochrome coaction. Science, 2001, 19: 487–490 10.1126/science.291.5503.487
Reed J W, Nagatani A, Elich T D, et al. Phytochrome A and phytochrome B have overlapping but distinct functions in Arabidopsis de velopment. Plant Physiol, 1994, 104: 1139–1149 1:CAS:528:DyaK2cXjtF2mtLw%3D, 12232154
Mockler T, Yang H, Yu X, et al. Regulation of photoperiodic flowering by Arabidopsis photoreceptors. Proc Natl Acad Sci USA, 2003, 100: 2140–2145 1:CAS:528:DC%2BD3sXhsFGrt7w%3D, 10.1073/pnas.0437826100, 12578985
Mockler T C, Guo H, Yang H, et al. Antagonistic actions of Arabidopsis cryptochromes and phytochrome B in the regulation of floral induction. Development, 1999, 126: 2073–2082 1:CAS:528:DyaK1MXjvVanu7g%3D, 10207133
Yu X, Shalitin D, Liu X, et al. De-repression of the NC80 motif is critical for the photoactivation of Arabidopsis CRY2. Proc Natl Acad Sci USA, 2007, 104: 7289–7294 1:CAS:528:DC%2BD2sXls1OgtLg%3D, 10.1073/pnas.0701912104, 17438275
Barnes S A, Nishizawa N K, Quaggio R B, et al. Far-red light blocks greening of Arabidopsis seedlings via a phytochrome A-mediated change in plastid development. Plant Cell, 1996, 8: 601–615 1:CAS:528:DyaK28Xis1Grtrw%3D, 10.1105/tpc.8.4.601, 8624438
Boylan M T, Quail P H. Phytochrome A overexpression inhibits hypocotyl elongation in transgenic Arabidopsis. Proc Natl Acad Sci USA, 1991, 88: 10806–10810 1:CAS:528:DyaK38XlsFOjsw%3D%3D, 10.1073/pnas.88.23.10806, 11607244
Parks B M, Quail P H. hy8, a new class of Arabidopsis long hypocotyl mutants deficient in functional phytochrome A. Plant Cell, 1993, 5: 39–48 1:CAS:528:DyaK3sXhs1GgtL4%3D, 10.1105/tpc.5.1.39, 8439743
McCormac A C, Terry M J. Loss of nuclear gene expression during the phytochrome A-mediated far-red block of greening response. Plant Physiol, 2002, 130: 402–414 1:CAS:528:DC%2BD38XntFOrt7c%3D, 10.1104/pp.003806, 12226519
Armstrong G A, Runge S, Frick G, et al. Identification of NADPH:protochlorophyllide oxidoreductases A and B: a branched pathway for light-dependent chlorophyll biosynthesis in Arabidopsis thaliana. Plant Physiol, 1995, 108: 1505–1517 1:CAS:528:DyaK2MXnsVagsb8%3D, 10.1104/pp.108.4.1505, 7659751
Holtorf H, Reinbothe S, Reinbothe C, et al. Two routes of chlorophyllide synthesis that are differentially regulated by light in barley (Hordeum vulgare L.). Proc Natl Acad Sci USA, 1995, 92: 3254–3258 1:CAS:528:DyaK2MXltFSntLw%3D, 10.1073/pnas.92.8.3254, 7724548
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Qin, Y., Guo, M., Li, X. et al. Stress responsive gene CIPK14 is involved in phytochrome A-mediated far-red light inhibition of greening in Arabidopsis. Sci. China Life Sci. 53, 1307–1314 (2010). https://doi.org/10.1007/s11427-010-4078-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11427-010-4078-1