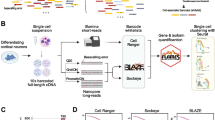
Abstract
Single-cell RNA sequencing (scRNA-seq) has become one of the most powerful tools to understand the heterogeneity of biological systems. While barcoding strategies have revolutionized the field of high-throughput scRNA-seq, it is still challenging to achieve highly efficient, direct and universal cell barcoding with cost-effectiveness and minimal sample loss. Herein, a single micro-particle dispenser approach for rapid single barcode bead/cell manipulation and pairing, enabling highly efficient cell barcoding for scRNA-seq (Dispen-Seq) was developed. Notably, Dispen-Seq provides a versatile platform which can enrich cell subgroups of interest while unlimited by input sample amounts, and can respond to changes in sample composition with high resolution and reproducibility. It is anticipated that Dispen-Seq will increase the scope of scRNA-seq from academic research to practical applications.

Similar content being viewed by others
References
Lu Y, Xue Q, Eisele MR, Sulistijo ES, Brower K, Han L, Amir ED, Peer D, Millerjensen K, Fan R. Proc Natl Acad Sci USA, 2015, 112: 201416756
Xu S, Ma W, Bai Y, Liu H. J Am Chem Soc, 2019, 141: 72–75
Fu Y, Chen H, Liu L, Huang Y. Anal Chem, 2016, 88: 10795–10799
Labib M, Mohamadi RM, Poudineh M, Ahmed SU, Ivanov I, Huang CL, Moosavi M, Sargent EH, Kelley SO. Nat Chem, 2018, 10: 489–495
Comi TJ, Do TD, Rubakhin SS, Sweedler JV. J Am Chem Soc, 2017, 139: 3920–3929
Krone KM, Warias R, Ritter C, Li A, Acevedo-Rocha CG, Reetz MT, Belder D. J Am Chem Soc, 2016, 138: 2102–2105
Xue M, Wei W, Su Y, Johnson D, Heath JR. J Am Chem Soc, 2016, 138: 3085–3093
Zhang Y, Wu M, Han X, Wang P, Qin L. Angew Chem Int Ed, 2015, 54: 10838–10842
Li J, Xun K, Pei K, Liu X, Peng X, Du Y, Qiu L, Tan W. J Am Chem Soc, 2019, 141: 18013–18020
Klein AM, Mazutis L, Akartuna I, Tallapragada N, Veres A, Li V, Peshkin L, Weitz DA, Kirschner MW. Cell, 2015, 161: 1187–1201
Zhang P, Han X, Yao J, Shao N, Zhang K, Zhou Y, Zu Y, Wang B, Qin L. Angew Chem Int Ed, 2019, 58: 13700–13705
Chattopadhyay PK, Chiu DT. Lab Chip, 2019, 19: 3573–3574
van Galen P, Hovestadt V, Wadsworth II MH, Hughes TK, Griffin GK, Battaglia S, Verga JA, Stephansky J, Pastika TJ, Lombardi Story J, Pinkus GS, Pozdnyakova O, Galinsky I, Stone RM, Graubert TA, Shalek AK, Aster JC, Lane AA, Bernstein BE. Cell, 2019, 176: 1265–1281.e24
Yun J, Zheng X, Xu P, Zheng X, Xu J, Cao C, Fu Y, Xu B, Dai X, Wang Y, Liu H, Yi Q, Zhu Y, Wang J, Wang L, Dong Z, Huang L, Huang Y, Du W. Small, 2020, 16: 1903739
Xu X, Wang J, Wu L, Guo J, Song Y, Tian T, Wang W, Zhu Z, Yang C. Small, 2020, 16: 1903905
Hanson WM, Chen Z, Jackson LK, Attaf M, Sewell AK, Heemstra JM, Phillips JD. J Am Chem Soc, 2016, 138: 11073–11076
Wu AR, Wang J, Streets AM, Huang Y. Annu Rev Anal Chem, 2017, 10: 439–462
Song Y, Xu X, Wang W, Tian T, Zhu Z, Yang C. Analyst, 2019, 144: 3172–3189
Rakszewska A, Stolper RJ, Kolasa AB, Piruska A, Huck WTS. Angew Chem Int Ed, 2016, 55: 6698–6701
Rosenberg AB, Roco CM, Muscat RA, Kuchina A, Sample P, Yao Z, Graybuck LT, Peeler DJ, Mukherjee S, Chen W, Pun SH, Sellers DL, Tasic B, Seelig G. Science, 2018, 360: 176–182
Jaitin DA, Kenigsberg E, Keren-Shaul H, Elefant N, Paul F, Zaretsky I, Mildner A, Cohen N, Jung S, Tanay A, Amit I. Science, 2014, 343: 776–779
Cao J, Spielmann M, Qiu X, Huang X, Ibrahim DM, Hill AJ, Zhang F, Mundlos S, Christiansen L, Steemers FJ, Trapnell C, Shendure J. Nature, 2019, 566: 496–502
Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, Trombetta JJ, Weitz DA, Sanes JR, Shalek AK, Regev A, McCarroll SA. Cell, 2015, 161: 1202–1214
Gierahn TM, Wadsworth Ii MH, Hughes TK, Bryson BD, Butler A, Satija R, Fortune S, Love JC, Shalek AK. Nat Methods, 2017, 14: 395–398
Zheng GXY, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, Ziraldo SB, Wheeler TD, McDermott GP, Zhu J, Gregory MT, Shuga J, Montesclaros L, Underwood JG, Masquelier DA, Nishimura SY, Schnall-Levin M, Wyatt PW, Hindson CM, Bharadwaj R, Wong A, Ness KD, Beppu LW, Deeg HJ, McFarland C, Loeb KR, Valente WJ, Ericson NG, Stevens EA, Radich JP, Mikkelsen TS, Hindson BJ, Bielas JH. Nat Commun, 2017, 8: 14049
Fan HC, Fu GK, Fodor SPA. Science, 2015, 347: 1258367
Dura B, Choi JY, Zhang K, Damsky W, Thakral D, Bosenberg M, Craft J, Fan R. Nucl Acids Res, 2018, 47: e16
Rouhanifard SH, Mellis IA, Dunagin M, Bayatpour S, Jiang CL, Dardani I, Symmons O, Emert B, Torre E, Cote A, Sullivan A, Stamatoyannopoulos JA, Raj A. Nat Biotechnol, 2019, 37: 84–89
Keren-Shaul H, Kenigsberg E, Jaitin DA, David E, Paul F, Tanay A, Amit I. Nat Protoc, 2019, 14: 1841–1862
Papalexi E, Satija R. Nat Rev Immunol, 2017, 18: 35–45
Whitfield ML, Sherlock G, Saldanha AJ, Murray JI, Botstein D. Mol Biol Cell, 2002, 13: 1977–2000
Zhang K, Gao M, Chong Z, Li Y, Han X, Chen R, Qin L. Lab Chip, 2016, 16: 4742–4748
Zhang K, Han X, Li Y, Li SY, Zu Y, Wang Z, Qin L. J Am Chem Soc, 2014, 136: 10858–10861
Lawrenz A, Nason F, Cooper-White JJ. Biomicrofluidics, 2012, 6: 024112
Cao J, Cusanovich DA, Ramani V, Aghamirzaie D, Pliner HA, Hill AJ, Daza RM, McFaline-Figueroa JL, Packer JS, Christiansen L, Steemers FJ, Adey AC, Trapnell C, Shendure J. Science, 2018, 361: 1380–1385
Acknowledgements
This work was supported by the National Key R&D Program of China (2018YFC1602900), the National Natural Science Foundation of China (21927806, 21735004, 222022409, 21874089, 21705024, 21775128) and the Program for Changjiang Scholars and Innovative Research Team in University (IRT13036).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest The authors declare no conflict of interest.
Supporting Information
11426_2020_9925_MOESM1_ESM.docx
Dispen-Seq: A Single-Microparticle Dispenser Based Strategy Towards Flexible Cell Barcoding for Single-Cell RNA Sequencing
Rights and permissions
About this article
Cite this article
Tian, T., Chen, Y., Bi, Y. et al. Dispen-Seq: a single-microparticle dispenser based strategy towards flexible cell barcoding for single-cell RNA sequencing. Sci. China Chem. 64, 650–659 (2021). https://doi.org/10.1007/s11426-020-9925-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11426-020-9925-8