Abstract
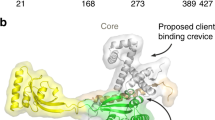
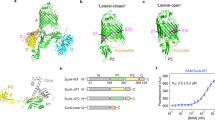
SurA is the major chaperone of outer membrane proteins (OMPs) in the periplasm. The molecular mechanism when SurA performs its chaperoning function is still unclear. Here, a purification-after-crosslinking (PAC) procedure was combined with single-molecule fluorescence resonance energy transfer (smFRET) to probe the conformations of SurA and OmpC in their complex. We found that SurA in the free state rearranges itself based on the crystal structure, except that the P2 domain moves towards the core domain with two major positions, forming a clamp-like conformation to accommodate OmpC. The obvious rearrangement of the P2 domain of SurA helps SurA to hold OmpC. OmpC attaches to SurA randomly and has the propensity to be near the middle part of the crevice. The noncollapsed and disordered conformations of OMPs provided by the OMPs·SurA complex are important to the subsequent delivery and folding process.
Similar content being viewed by others
References
Schulz GE. Curr Opin Struct Biol, 2000, 10: 443–447
Breyton C, Haase W, Rapoport TA, Kühlbrandt W, Collinson I. Nature, 2002, 418: 662–665
Van den Berg B, Clemons WM, Collinson I, Modis Y, Hartmann E, Harrison SC, Rapoport TA. Nature, 2004, 427: 36–44
Rapoport TA. Nature, 2007, 450: 663–669
Driessen AJM, Nouwen N. Annu Rev Biochem, 2008, 77: 643–667
Sklar JG, Wu T, Kahne D, Silhavy TJ. Genes Dev, 2007, 21: 2473–2484
Knowles TJ, Scott-Tucker A, Overduin M, Henderson IR. Nat Rev Microbiol, 2009, 7: 206–214
Lyu ZX, Zhao XS. Biochem Soc Trans, 2015, 43: 133–138
Noinaj N, Gumbart JC, Buchanan SK. Nat Rev Microbiol, 2017, 15: 197–204
Lazar SW, Kolter R. J Bacteriol, 1996, 178: 1770–1773
Rouvière PE, Gross CA. Genes Dev, 1996, 10: 3170–3182
Behrens S, Maier R, de CH, Schmid FX, Gross CA. EMBO J, 2001, 20: 285–294
O’Neil PK, Rollauer SE, Noinaj N, Buchanan SK. Biochemistry, 2015, 54: 6303–6311
Bitto E, McKay DB. Structure, 2002, 10: 1489–1498
Soltes GR, Schwalm J, Ricci DP, Silhavy TJ. J Bacteriol, 2016, 198: 921–929
Liu CP, Zhou QM, Fan DJ, Zhou JM. Int J Biochem Cell Biol, 2010, 42: 890–901
Bitto E, McKay DB. J Biol Chem, 2003, 278: 49316–49322
Bitto E, McKay DB. FEBS Lett, 2004, 568: 94–98
Hennecke G, Nolte J, Volkmer-Engert R, Schneider-Mergener J, Behrens S. J Biol Chem, 2005, 280: 23540–23548
Xu X, Wang S, Hu YX, McKay DB. J Mol Biol, 2007, 373: 367–381
Wu S, Ge X, Lv Z, Zhi Z, Chang Z, Zhao XS. Biochem J, 2011, 438: 505–511
Zhong M, Ferrell B, Lu W, Chai Q, Wei Y. J Bacteriol, 2013, 195: 1061–1067
Li G, He C, Bu P, Bi H, Pan S, Sun R, Zhao XS. ACS Chem Biol, 2018, 13: 1082–1089
Krainer G, Keller S, Schlierf M. Curr Opin Struct Biol, 2019, 58: 124–137
He S, Yang C, Peng S, Chen C, Zhao XS. RSC Adv, 2019, 9: 14745–14749
Baslé A, Rummel G, Storici P, Rosenbusch JP, Schirmer T. J Mol Biol, 2006, 362: 933–942
Thoma J, Burmann BM, Hiller S, Müller DJ. Nat Struct Mol Biol, 2015, 22: 795–802
Sinz A, Arlt C, Chorev D, Sharon M. Protein Sci, 2015, 24: 1193–1209
Han L, Zheng J, Wang Y, Yang X, Liu Y, Sun C, Cao B, Zhou H, Ni D, Lou J, Zhao Y, Huang Y. Nat Struct Mol Biol, 2016, 23: 192–196
Wang Y, Wang R, Jin F, Liu Y, Yu J, Fu X, Chang Z. J Biol Chem, 2016, 291: 16720–16729
Mas G, Hiller S. FEMS MicroBiol Lett, 2018, 365
Chai Q, Ferrell B, Zhong M, Zhang X, Ye C, Wei Y. Protein Eng Des Sel, 2014, 27: 111–116
Struyvé M, Moons M, Tommassen J. J Mol Biol, 1991, 218: 141–148
Robert V, Volokhina EB, Senf F, Bos MP, Van Gelder P, Tommassen J. PLoS Biol, 2006, 4: e377
Dunker AK, Obradovic Z. Nat Biotechnol, 2001, 19: 805–806
Dunker AK, Lawson JD, Brown CJ, Williams RM, Romero P, Oh JS, Oldfield CJ, Campen AM, Ratliff CM, Hipps KW, Ausio J, Nissen MS, Reeves R, Kang CH, Kissinger CR, Bailey RW, Griswold MD, Chiu W, Garner EC, Obradovic Z. J Mol Graphics Model, 2001, 19: 26–59
Moon CP, Zaccai NR, Fleming PJ, Gessmann D, Fleming KG. Proc Natl Acad Sci USA, 2013, 110: 4285–4290
Bu PX, He CH, Zhao XS. Acta Phys-Chim Sin, 2019, 35: 546–554
Hagan CL, Westwood DB, Kahne D. Biochemistry, 2013, 52: 6108–6113
Hagan CL, Kahne D. Biochemistry, 2011, 50: 7444–7446
Mogensen JE, Otzen DE. Mol Microbiol, 2005, 57: 326–346
Acknowledgements
This work was supported by the National Natural Science Foundation of China (21233002, 21521003) and the National Key Basic Research Special Foundation of China (2012CB917304). The measurements of CD spectra were performed at the Analytical Instrumentation Center of Peking University (PKUAIC). We acknowledge the assistance of Dr. Jiang Zhou from PKUAIC.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest The authors declare that they have no conflict of interest.
Supporting Information
11426_2020_9758_MOESM1_ESM.pdf
Molecular Mechanism of SurA’s Chaperoning Function to Outer Membrane Proteins Revealed by Purification-after-Crosslinking Single-Molecule FRET
Rights and permissions
About this article
Cite this article
He, C., Pan, S., Li, G. et al. Molecular mechanism of SurA’s chaperoning function to outer membrane proteins revealed by purification-after-crosslinking single-molecule FRET. Sci. China Chem. 63, 1142–1152 (2020). https://doi.org/10.1007/s11426-020-9758-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11426-020-9758-2