Abstract
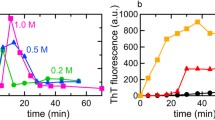
Our dynamic laser light scattering (LLS) study shows that the current widely used protocols of dissolving amyloidogenic protein/peptide do not really result in a true solution; namely, there always exist a trace amount of interchain aggregates, which greatly affect the association kinetics, partially explaining why different kinetics were reported even for a solution with identical protein and solvent. Recently, using a combination of the conventional dissolution procedure and our newly developed ultra-filtration method, we have developed a novel protocol to prepare a true solution of amyloidogenic protein/peptide without any interchain aggregates. The resultant solutions remain in their monomeric state for at least one week, which is vitally important for further study of the very initial stage of the interchain association under the physiological conditions because more and more evidence suggests that it is those small oligomers rather than large fabric aggregates that are cytotoxic. In addition, this study shows that combining static and dynamic LLS can lead to more physical and microscopic information about the protein association instead of only the size distribution.
Similar content being viewed by others
References
Selkoe DJ. Folding proteins in fatal ways. Nature, 2003, 426: 900–904
Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem, 2006, 75: 333–366
Kayed R, Head E, Thompson JL, McIntire TM, Milton SC, Cotman CW, Glabe CG. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science, 2003, 300: 486–489
Shankar GM, Li S, Mehta TH, Garcia-Munoz A, Shepardson NE, Smith I, Brett FM, Farrell MA, Rowan MJ, Lemere CA, Regan CM, Walsh DM, Sabatini BL, Selkoe DJ. Amyloid-beta protein dimers isolated directly from Alzheimer's brains impair synaptic plasticity and memory. Nat Med, 2008, 14: 837–842
Palhano FL, Rocha CB, Bernardino A, Weissmuller G, Masuda CA, Montero-Lomelí M, Gomes AM, Chien P, Fernandes PM, Foguel DA. fluorescent mutant of the NM domain of the yeast prion Sup35 provides insight into fibril formation and stability. Biochemistry, 2009, 48: 6811–6823
Glover JR, Kowal AS, Schirmer EC, Patino MM, Liu JJ, Lindquist S. Self-seeded fibers formed by Sup35, the protein determinant of [PSI+], a heritable prion-like factor of S. cerevisiae. Cell, 1997, 89: 811–819
Hess S, Lindquist SL, Scheibel T. Alternative assembly pathways of the amyloidogenic yeast prion determinant Sup35-NM. Embo Reports, 2007, 8: 1196–1201
Giuffrida ML, Caraci F, Pignataro B, Cataldo S, De Bona P, Bruno V, Molinaro G, Pappalardo G, Messina A, Palmigiano A, Garozzo D, Nicoletti F, Rizzarelli E, Copani A. Beta-amyloid monomers are neuroprotective. J Neurosci, 2009, 29: 10582–10587
Georgalis Y, Starikov EB, Hollenbach B, Lurz R, Scherzinger E, Saenger W, Lehrach H, Wanker EE. Huntingtin aggregation monitored by dynamic light scattering. Proc Natl Acad Sci USA, 1998, 95: 6118–6121
Chen SM, Wetzel R. Solubilization and disaggregation of polyglutamine peptides. Protein Sci, 2001, 10: 887–891
Scheibel T, Lindquist SL. The role of conformational flexibility in prion propagation and maintenance for Sup35p. Nat Struct Biol, 2001, 8: 958–962
Cummings CJ, Zoghbi HY. Trinucleotide repeats: mechanisms and pathophysiology. Annu Rev Genomics Hum Genet, 2000, 1: 281–328
Walker FO. Huntington's disease. Lancet, 2007, 369: 218–228
Chen CY, Rojanatavorn K, Clark AC, Shih JC. Characterization and enzymatic degradation of Sup35NM, a yeast prion-like protein. Protein Sci, 2005, 14: 2228–2235
Walsh DM, Selkoe DJ. A beta oligomers-a decade of discovery. J Neurochem, 2007, 101: 1172–1184
Chen QJ, Zhao H, Ming T, Wang JF, Wu C. Nanopore extrusion-induced transition from spherical to cylindrical block copolymer micelles. J Am Chem Soc, 2009, 131: 16650–16651
Ge H, Jin F, Li JF, Wu C. How much force is needed to stretch a coiled chain in solution? Macromolecules, 2009, 42: 4400–4402
Ge H, Wu C. Separation of linear and star chains by a nanopore. Macromolecules, 2010, 43: 8711–8713
Foguel D, Palhano FL, Rocha CB, Bernardino A, Weissmuller G, Masuda CA, Montero-Lomeli M, Gomes AM, Chien P, Fernandes PMB. A fluorescent mutant of the NM domain of the yeast prion Sup35 provides insight into fibril formation and stability. Biochemistry, 2009, 48: 6811–6823
Tanaka M, Ohhashi Y, Ito K, Toyama BH, Weissman JS. Differences in prion strain conformations result from non-native interactions in a nucleus. Nat Chem Bio, 2010, 6: 225–230
Wu C, Xia KQ. Incorporation of a differential refractometer into a laser light-scattering spectrometer. Rev Sci Instrum, 1994, 65: 587–590
LeVine H. Quantification of beta-sheet amyloid fibril structures with thioflavin T. In Amyloid, Prions, and Other Protein Aggregates. Academic Press Inc, 1999, 309: 274
Colby DW, Zhang Q, Wang S, Groth D, Legname G, Riesner D, Prusiner SB. Prion detection by an amyloid seeding assay. Proc Natl Acad Sci USA, 2007, 105: 1774–11774
Garcia-Martin F, Quintanar-Audelo M, Garcia-Ramos Y, Cruz LJ, Gravel C, Furic R, Côté S, Tulla-Puche J, Albericio F. ChemMatrix, a poly(ethylene glycol)-based support for the solid-phase synthesis of complex peptides. J Comb Chem, 2006, 8: 213–22
Author information
Authors and Affiliations
Corresponding author
Additional information
Contributed equally to this work.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Diao, S., Zhao, H., Wang, W. et al. Preparation of true solutions of monomeric amyloidogenic protein/peptide: A critical prerequisite for aggregation kinetic study. Sci. China Chem. 55, 118–124 (2012). https://doi.org/10.1007/s11426-011-4446-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11426-011-4446-0