Abstract
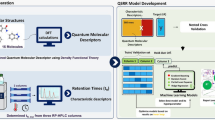
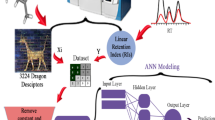
An integrated approach is proposed to predict the chromatographic retention time of oligonucleotides based on quantitative structure-retention relationships (QSRR) models. First, the primary base sequences of oligonucleotides are translated into vectors based on scores of generalized base properties (SGBP), involving physicochemical, quantum chemical, topological, spatial structural properties, etc.; thereafter, the sequence data are transformed into a uniform matrix by auto cross covariance (ACC). ACC accounts for the interactions between bases at a certain distance apart in an oligonucleotide sequence; hence, this method adequately takes the neighboring effect into account. Then, a genetic algorithm is used to select the variables related to chromatographic retention behavior of oligonucleotides. Finally, a support vector machine is used to develop QSRR models to predict chromatographic retention behavior. The whole dataset is divided into pairs of training sets and test sets with different proportions; as a result, it has been found that the QSRR models using more than 26 training samples have an appropriate external power, and can accurately represent the relationship between the features of sequences and structures, and the retention times. The results indicate that the SGBP-ACC approach is a useful structural representation method in QSRR of oligonucleotides due to its many advantages such as plentiful structural information, easy manipulation and high characterization competence. Moreover, the method can further be applied to predict chromatographic retention behavior of oligonucleotides.
Similar content being viewed by others
References
Huber CG. Micropellicular stationary phases for high-performance liquid chromatography of double-stranded DNA. J Chromatogr A, 1998, 806: 1–28
Gilar M, Fountain KJ, Budman Y, Neue UD, Yardley KR, Rainville PD, Russell RJ, Gebler JC. Ion-pair reversed-phase high-performance liquid chromatography analysis of oligonucleotides: retention prediction. J Chromatogr A, 2002, 958: 167–182
Kaliszan R, Fork H. The relationship between the RM values and the connectivity indices for pyrazine carbothioamide derivatives. Chromatographia, 1977, 10: 346–355
Kaliszan R. Correlation between the retention indices and the connectivity indices of alcohols and methyl esters with complex cyclic structure. Chromatographia, 1977, 10: 529–540
Michotte Y, Massart DL. Molecular connectivity and retention indexes. J Pharm Sci, 1977, 66: 1630–1632
Héberger K. Quantitative structure-(chromatographic) retention relationships. J Chromatogr A, 2007, 1158: 273–305
Put R, Heyden YV. Review on modelling aspects in reversed-phase liquid chromatographic quantitative structure-retention relationships. Anal Chim Acta, 2007, 602:164–172
Kaliszan R. QSRR: Quantitative structure-(chromatographic) retention relationships. Chem Rev, 2007, 107: 3212–3246
Put R, Daszykowski M, Baczek T, Vander Heyden Y. Retention prediction of peptides based on uninformative variable elimination by partial least squares. J Proteome Res, 2006, 5: 1618–1625
Baczek T, Wiczling P, Marszall M, Heyden YV. Kaliszan R. Prediction of peptide retention at different HPLC conditions from multiple linear regression models. J Proteome Res, 2005, 4: 555–563
Bodzioch K, Baczek T, Kaliszan R, Vander Heyden Y. The molecular descriptor log SumAA and its alternatives in QSRR models to predict the retention of peptides. J Pharm Biomed Anal, 2009, 50: 563–569.
Ladiwala A, Xia F, Luo Q, Breneman CN, Cramer SM. Investigation of protein retention and selectivity in HIC systems using quantitative structure retention relationship models. Biotechnol Bioeng, 2006, 93: 836–850
Kohlbacher O, Quinten S, Sturm M, Mayr B, Huber C. Structureactivity relationships in chromatography: Retention prediction of oligonucleotides with support vector regression. Angew Chem Int Ed, 2006, 45: 7009–7012
Harju M, Andersson PL, Haglund P, Tysklind M. Multivariate physicochemical characterisation and quantitative structure-property relationship modelling of polybrominated diphenyl ethers. Chemosphere, 2002, 47: 375–384
Bucinski A, Wnuk M, Goryński K, Giza A, Kochańczyk J, Nowaczyk A, Bączek T, Nasal A. Artificial neural networks analysis used to evaluate the molecular interactions between selected drugs and human α1-acid glycoprotein. J Pharm Biomed Anal, 2009, 50: 591–596
Can H, Dimoglo A, Kovalishyn V. Application of artificial neural networks for the prediction of sulfur polycyclic aromatic compounds retention indices. J Mol Struct (THEOCHEM), 2005, 723: 183–188
Yang C, Zhong C. Chirality factors and their application to QSAR studies of chiral molecules. QSAR Comb Sci, 2005, 24: 1047–1055
Rybolt TR, Janeksela VE, Hooper DN, Thomas HE, Carrington NA, Williamson EJ. Predicting second gas-solid virial coefficients using calculated molecular properties on various carbon surfaces. J Colloid Interface Sci, 2004, 272: 35–45
Skrbic B, Onjia A. Prediction of the Lee retention indices of polycyclic aromatic hydrocarbons by artificial neural network. J Chromatogr A, 2006, 1108: 279–284
Liang GZ, Li ZL. Scores of generalized base properties for quantitative sequence-activity modelings for E.coli promoters based on support vector machine. J Mol Graph Model, 2007, 26: 269–281
Johnson RA, Wichern DW. Applied Multivariate Statistical Analysis. New Jersey: Prentice Hall, 2002
Nystrom A, Andersson PM, Lundstedt T. Multivariate data analysis of topographically modified α-melanotropin analogues using auto and cross auto covariances. Quant Struct-Act Relat, 2000, 19: 264–269
Leardi R, Lupianez A. Genetic algorithms applied to feature selection in PLS regression: How and when to use them. Chemolab, 1998, 41: 195–207
Vapnik V. Statistical Learning Theory. NewYork: Wiley-Interscience, 1998
Chou KC, Shen HB. Cell-PLoc: A package of Web servers for predicting subcellular localization of proteins in various organism. Nat Protoc, 2008, 3: 153–162
Li WJ, Wu JJ. The construction of RNA secondary structure prediction system. Progr Biochem Biophys, 1996, 23: 449–453
Zou HF, Zhang YK, Hong MF, Lu PC. Retention behavior of small peptides in reversed-phase high-performance liquid chromatography. Chin J Chromatogr, 1991, 9: 257–262
Huber CG, Oefner PJ, Bonn GK. High-performance liquid chromatographic separation of detritylated oligonucleotides on highly cross-linked poly-(styrene-divinylbenzene) particles. J Chromatogr, 1992, 599: 113–118
Dickman JM. Effects of sequence and structure in the separation of nucleic acids using ion pair reverse phase liquid chromatography. J Chromatogr A, 2005, 1076: 83–89
Mathews DH, Disney MD, Childs JL, Schroeder SJ, Zuker M, Turner DH. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc Natl Acad Sci USA, 2004, 101: 7287–7292
Brodsky LI, Ivanov VV, Kalaidzidis YL, Leontovich AM, Nikolaev VK, Feranchuk SI, Drachev VA. GeneBee-NET: internet-based server for analyzing biopolymers structure. Biochemistry, 1995, 60: 923–928
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Zhao, W., Liang, G., Chen, Y. et al. A new quantitative structure-retention relationship model for predicting chromatographic retention time of oligonucleotides. Sci. China Chem. 54, 1064–1071 (2011). https://doi.org/10.1007/s11426-011-4299-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11426-011-4299-6