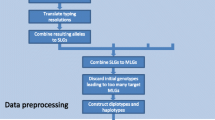
Abstract
This paper discusses the associations between traits and haplotypes based on FI (fluorescent intensity) data sets. We consider a clustering algorithm based on mixtures of t distributions to obtain all possible genotypes of each individual (i.e. “GenoSpectrum”). We then propose a likelihood-based approach that incorporates the genotyping uncertainty to assessing the associations between traits and haplotypes through a haplotype-based logistic regression model. Simulation studies show that our likelihood-based method can reduce the impact induced by genotyping errors.
Similar content being viewed by others
References
Epstein, M. P., Satten, G. A., Inference on haplotype effects in case-control studies using unphased genotype data, Am. J. Hum. Genet., 2003, 73: 1316–1329.
Akey, J. M., Zhang, K., Xiong, M. et al., The effect that genotyping errors have on the robustness of common linkage-disequilibrium measures, Am. J. Hum. Genet., 2001, 68: 1447–1456.
Fallin, D., Cohen, A., Essioux, L. et al., Genetic analysis of case/control data using estimated haplotype frequencies: application to APOE locus variation and Alzheimer's disease, Genome Res., 2001, 11: 143–151.
Zaykin, D. V., Westfall, P. H., Young, S. S. et al., Testing association of statistically inferred haplotypes with discrete and continuous traits in samples of unrelated individuals, Hum. Hered., 2002, 50: 133–139.
Michalatos-Beloin, S., Tishkoff, S. A., Bentley, K. L. et al., Molecular haplotyping of genetic markers 10kb apart by allele-sqecific long-rang PCR, Nucleic. Acids Res., 1996, 24: 4841–4843.
Yan, H., Papadopoulos, N., Marra, G. et al., Conversion of diploidy to haploidy, Nature, 2000, 403: 723–724.
Douglas, J. A., Boehnke, M., Gillanders, E., et al., Experimentally-derived haplotypes substantially increase the efficiency of linkage disequilibrium studies, Nat. Genet., 2001, 28: 361–364.
Eitan, Y., Kashi, Y., Direct micro-haplotyping by multiple double PCR amplifications of specific alleles (MD-PASA), Nucleic. Acids Res., 2002, 30: e62.
Clark, A. G., Inference of haplotypes from PCR-amplified samples of diploid populations, Mol. Biol. Evol., 1990, 7: 111–122.
Excoffier, L., Slatkin, M., Maximum-likelihood estimation of molecular haplotype frequencies in a diploid population, Mol. Biol. Evol., 1995, 12: 921–927.
Hawley, M. E., Kidd, K. K., HAPLO: A program using the EM algorithm to estimate the frequencies of multi-site haplotypes. J. Hered., 1995, 86: 409–411.
Long, J. C., Williams, R. C., Urbanek, M., An E-M algorithm and testing strategy for multiple-locus haplotypes, Am. J. Hum. Genet., 1995, 56: 799–810.
Stephens, M., Smith, N. J., Donnelly, P., A new statistical method for haplotype reconstruction from population data, Am. J. Hum. Genet., 2001, 68: 978–989.
Lin, S., Cutler, D. J., Zwick, M. E. et al., Haplotype Inference in random population samples, Am. J. Hum. Genet., 2002, 71: 1129–1137.
Niu, T., Qin, Z. S., Xu, X. et al., Bayesian haplotype inference for multiple linked singe-nucleotide polymorphisms, Am. J. Hum. Genet., 2002, 70: 157–169.
Qin, Z. S., Niu, T., Liu, J. S., Partition-ligation-expectation-maximization algorithm for haplotype inference with singe-nucleotide polymorphisms, Am. J. Hum. Genet., 2002, 71: 1242–1247.
Stephens, M., Donnelly, P., A comparison of Bayesian methods for haplotype reconstruction from population genotype data, Am. J. Hum. Genet., 2003, 73: 1162–1169.
Zhao, J. H., Curtis, D., Sham, P. C., Model-free analysis and permutation tests for allelic association, Hum. Hered., 2000, 50: 133–139.
Schaid, C. J., Score test for association between traits and haplotypes when linkage phase is ambiguous, Am. J. Hum. Genet., 2002, 70: 425–434.
Zhao, L. P., Li, S. S., Khalid, N., A method for the assessment of disease associations with single-nucleotide polymorphism haplotypes and environmental variables in case-control studies, Am. J. Hum. Genet., 2003, 72: 1231–1250.
Stram, D. O., Haiman, C. A., Hirschhorn, J. H. et al., Choosing haplotype-tagging SNPs based on unphased genotype data form a preliminary sample of unrelated subjects with an example from the Multiethnic Cohort study, Hum. Hered., 2003a, 55: 27–36.
Kang, H., Qin, Z. S., Niu, T. et al., Incorporating genotyping uncertainty in haplotype inference for single-nucleotide polymorphisms, Am. J. Hum. Genet., 2004, 74: 495–510.
Sobel, E., Papp, J. C., Lange, K., Detection and integration of genotyping errors in statistical genetics, Am. J. Hum. Genet., 2002, 70: 496–508.
Kirk, K. M., Cardon, L. R., The impact of genotyping error on haplotype reconstruction and frequency estimation, Eur. J. Hum. Genet., 2002, 10: 616–622.
Buetow, K. H., Influence of aberrant observations on high resolution linkage analysis outcome, Am. J. Hum. Genet., 1991, 49: 985–994.
Goldstein, D. R., Zhao, H., Speed, T. P., The effects of genotyping errors and interference on estimation of genetics distance, Hum. Hered., 1997, 47: 86–100.
Douglas, J. A., Boehnke, M., Lange, K., A multipoint method for detecting genotyping errors and mutations in sibling-pair linkage data, Am. J. Hum. Genet., 2000, 66: 1287–1297.
Douglas, J. A., Skol, A. D., Boehnke, M., Probability of detection of genotyping errors and mutations as inheritance inconsistencies in nuclear-family data, Am. J. Hum. Genet., 2002, 70: 487–495.
Abecasis, G. R., Cherny, S. S., Cardon, L. R., The impact of genotyping error on family-based analysis of quantitative traits, Eur. J. Hum. Genet., 2001, 9: 130–134.
Wallenstein, S., Hodge, S. E., Weston, A., Logistic regression model for analyzing extended haplotype data, Genet. Epidemiol., 1998, 15: 173–181.
Dempster, A. P., Laird, N. M., Rubin, D. B., Maximum likelihood from incomplete data via EM algorithm, J. Roy. Stat. Soc. Ser. B, 1977, 39: 1–38.
Ibrahim, J. G., Incomplete data in generalized linear models, J. Am. Statist. Assoc., 1990, 85: 765–769.
Louis, T. A., Finding the observed information matrix when using the EM algorithm, J. Roy. Statist. Soc. B, 1982, 44: 226–233.
Horton, N. J., Laird, N. M., Maximum likelihood analysis of logistic regression models with incomplete covariate data and auxiliary information, Biometrics, 2001, 57: 34–42.
Schaid, C. J., Evaluating associations of haplotypes with traits, Genet. Epidemiol., 2004, 27: 348–364.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Zhu, W., Guo, J. A likelihood-based method for haplotype association studies of case-control data with genotyping uncertainty. SCI CHINA SER A 49, 130–144 (2006). https://doi.org/10.1007/s11425-005-0147-5
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/s11425-005-0147-5