Abstract
Frailty, a prevalent clinical syndrome in aging adults, is characterized by poor health outcomes, represented via a standardized frailty-phenotype (FP), and Frailty Index (FI). While the relevance of the syndrome is gaining awareness, much remains unclear about its underlying biology. Further elucidation of the genetic determinants and possible underlying mechanisms may help improve patients’ outcomes allowing healthy aging.
Genotype, clinical and demographic data of subjects (aged 60–73 years) from UK Biobank were utilized. FP was defined on Fried’s criteria. FI was calculated using electronic-health-records. Genome-wide-association-studies (GWAS) were conducted and polygenic-risk-scores (PRS) were calculated for both FP and FI. Functional analysis provided interpretations of underlying biology. Finally, machine-learning (ML) models were trained using clinical, demographic and PRS towards identifying frail from non-frail individuals.
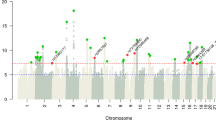
Thirty-one loci were significantly associated with FI accounting for 12% heritability. Seventeen of those were known associations for body-mass-index, coronary diseases, cholesterol-levels, and longevity, while the rest were novel. Significant genes CDKN2B and APOE, previously implicated in aging, were reported to be enriched in lipoprotein-particle-remodeling. Linkage-disequilibrium-regression identified specific regulation in limbic-system, associated with long-term memory and cognitive-function. XGboost was established as the best performing ML model with area-under-curve as 85%, sensitivity and specificity as 0.75 and 0.8, respectively.
This study provides novel insights into increased vulnerability and risk stratification of frailty syndrome via a multi-modal approach. The findings suggest frailty as a highly polygenic-trait, enriched in cholesterol-remodeling and metabolism and to be genetically associated with cognitive abilities. ML models utilizing FP and FI + PRS were established that identified frailty-syndrome patients with high accuracy.




Similar content being viewed by others
Data availability
Data supporting the results reported in the manuscript are obtained from UK Biobank (www.ukbiobank.ac.uk), a major biomedical database, approved under project # 54423.
References
W. H. Organization, "GHE: life expectancy and healthy life expectancy," Available online at: https://www.who.int/data/gho/data/themes/mortality-and-global-health-estimates/ghe-life-expectancy-and-healthy-life-expectancy, no. Accessed in August 2022, 2022.
van den Heuvel WJ, Olaroiu M. How important are health care expenditures for life expectancy? A comparative, European analysis. J Am Med Dir Assoc. 2017;18(3):276.e9-276.e12.
Jaba E, Balan CB, Robu I-B. The relationship between life expectancy at birth and health expenditures estimated by a cross-country and time-series analysis. Procedia Econ Finan. 2014;15:108–14.
Lubitz J, Beebe J, Baker C. Longevity and medicare expenditures. 1995;332(15):999–1003. https://doi.org/10.1056/nejm199504133321506.
Lubitz JD, Riley GF, Trends in Medicare payments in the last year of life. 1993;328 (15):1092–1096. https://doi.org/10.1056/nejm199304153281506.
Spillman BC, Lubitz J. The effect of longevity on spending for acute and long-term care. 2000;342(19):1409–1415. https://doi.org/10.1056/nejm200005113421906.
Collard RM, Boter H, Schoevers RA, Oude Voshaar RC. Prevalence of frailty in community-dwelling older persons: a systematic review. J Am Ger Soc. 2012;60(8):1487–92.
Eyigor S, et al. Frailty prevalence and related factors in the older adult—FrailTURK Project. Age. 2015;37:1–13.
O’Caoimh R, et al. Prevalence of frailty in 62 countries across the world: a systematic review and meta-analysis of population-level studies. Age and Ageing. 2021;50(1):96–104. https://doi.org/10.1093/ageing/afaa219. (in English).
Ma Y et al. The association between frailty and severe disease among COVID-19 patients aged over 60 years in China: a prospective cohort study. Bmc Med. 2020;18(1). Art no. 274, https://doi.org/10.1186/s12916-020-01761-0.
Welch C. Age and frailty are independently associated with increased COVID-19 mortality and increased care needs in survivors: results of an international multi-centre study. Age Ageing. 2021;50(3):617–30.
Murad K, Kitzman DW. Frailty and multiple comorbidities in the elderly patient with heart failure: implications for management. Heart Fail Rev. 2012;17(4–5):581–8. https://doi.org/10.1007/s10741-011-9258-y.
Mhaolain AMN, et al. Frailty, depression, and anxiety in later life. Int Psychogeriatr. 2012;24(8):1265–74. https://doi.org/10.1017/s1041610211002110.
Xue QL. The Frailty Syndrome: definition and natural history, (in English). Clin Geriatr Med. 2011;27(1):1. https://doi.org/10.1016/j.cger.2010.08.009.
Harmand MG-C, et al. Comparing the predictive value of three definitions of frailty: results from the three-city study. Arch Gerontol Geriatr. 2017;72:153–63.
Cesari M, Gambassi G, Abellan van Kan G, Vellas B. The frailty phenotype and the frailty index: different instruments for different purposes. Age Ageing. 2014;43(1):10–2.
Mitnitski AB, Mogilner AJ, Rockwood K. Accumulation of deficits as a proxy measure of aging, (in eng). Sci World J. 2001;1:323–36. https://doi.org/10.1100/tsw.2001.58.
Gale CR, Cooper C, Aihie Sayer A. Prevalence of frailty and disability: findings from the English Longitudinal Study of Ageing. Age Ageing. 2014;44(1):162–5. https://doi.org/10.1093/ageing/afu148.
O’Caoimh R, et al. Prevalence of frailty in 62 countries across the world: a systematic review and meta-analysis of population-level studies. Age Ageing. 2021;50(1):96–104.
Hoogendijk EO, Afilalo J, Ensrud KE, Kowal P, Onder G, Fried LP. Frailty: implications for clinical practice and public health, (in English). Lancet, Article. 2019;394(10206):1365–75. https://doi.org/10.1016/s0140-6736(19)31786-6.
Won CW. Diagnosis and management of frailty in primary health care. Korean J Fam Med. 2020;41(4):207.
Livshits G, et al. Shared genetic influence on frailty and chronic widespread pain: a study from TwinsUK. Age Ageing. 2018;47(1):119–25. https://doi.org/10.1093/ageing/afx122.
Young ACM, Glaser K, Spector TD, Steves CJ. The identification of hereditary and environmental determinants of frailty in a cohort of UK Twins. Twin Res Hum Genet. 2016;19(6):600–9. https://doi.org/10.1017/thg.2016.72.
Kim S, Welsh DA, Cherry KE, Myers L, Jazwinski SM. Association of healthy aging with parental longevity, (in eng). Age (Dordr). 2013;35(5):1975–82. https://doi.org/10.1007/s11357-012-9472-0.
Atkins JL et al. A genome-wide association study of the frailty index highlights brain pathways in ageing. 2021;20(9):e13459. https://doi.org/10.1111/acel.13459.
Ravindrarajah R, Hazra NC, Charlton J, Jackson SHD, Dregan A, Gulliford MC. Incidence and mortality of fractures by frailty level over 80 years of age: cohort study using UK electronic health records. 2018;8(1):e018836. https://doi.org/10.1136/bmjopen-2017-018836.
Petermann-Rocha F, et al. Comparison of two different frailty measurements and risk of hospitalisation or death from COVID-19: findings from UK Biobank. BMC Med. 2020;18(1):355. https://doi.org/10.1186/s12916-020-01822-4.
Martin GP et al. Do frailty measures improve prediction of mortality and morbidity following transcatheter aortic valve implantation? An analysis of the UK TAVI registry. 2018;8(6):e022543. https://doi.org/10.1136/bmjopen-2018-022543.
Parmar KL, Law J, Carter B, et al. Frailty in older patients undergoing emergency laparotomy: results from the uk observational emergency laparotomy and frailty (ELF) study. Ann Surg. 2021;273(4):709–718. https://doi.org/10.1097/SLA.0000000000003402
Petermann-Rocha F, et al. Associations between physical frailty and dementia incidence: a prospective study from UK Biobank. Lancet Health Longev. 2020;1(2):e58–68. https://doi.org/10.1016/S2666-7568(20)30007-6.
Bycroft C, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562(7726):203–9. https://doi.org/10.1038/s41586-018-0579-z.
Ye Y, Noche RB, Szejko N, et al. A genome-wide association study of frailty identifies significant genetic correlation with neuropsychiatric, cardiovascular, and inflammation pathways [published online ahead of print, 2023 Mar 16]. Geroscience. 2023. https://doi.org/10.1007/s11357-023-00771-z
Atkins JL, et al. A genome-wide association study of the frailty index highlights brain pathways in ageing. Aging Cell. 2021;20(9):e13459.
W. H. Organization. Ageing overview. https://www.who.int/health-topics/ageing#tab=tab_1.
Sudlow C, et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12(3):e1001779.
Searle SD, Mitnitski A, Gahbauer EA, Gill TM, Rockwood K. A standard procedure for creating a frailty index. BMC Geriatr. 2008;8(1):24. https://doi.org/10.1186/1471-2318-8-24.
Howlett SE, Rockwood MRH, Mitnitski A, Rockwood K. Standard laboratory tests to identify older adults at increased risk of death. BMC Med. 2014;12(1):171. https://doi.org/10.1186/s12916-014-0171-9.
Pajewski NM, Lenoir K, Wells BJ, Williamson JD, Callahan KE. Frailty screening using the electronic health record within a Medicare accountable care organization. J Gerontol: Series A. 2019;74(11):1771–7.
Fried LP, et al. Frailty in older adults: evidence for a phenotype, (in English). J Gerontol Ser A-Biol Sci Med Sci. 2001;56(3):M146–56. https://doi.org/10.1093/gerona/56.3.M146.
de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLOS Comput Biol. 2015;11(4):e1004219. https://doi.org/10.1371/journal.pcbi.1004219.
Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nature Commun. 2017;8(1):1826. https://doi.org/10.1038/s41467-017-01261-5.
Finucane HK, et al. Heritability enrichment of specifically expressed genes identifies disease-relevant tissues and cell types, (in eng). Nat Genet. 2018;50(4):621–9. https://doi.org/10.1038/s41588-018-0081-4.
McGeary SE, et al. The biochemical basis of microRNA targeting efficacy. Science. 2019;366(6472):eaav1741.
Karagkouni D, et al. DIANA-TarBase v8: a decade-long collection of experimentally supported miRNA–gene interactions. Nucleic Acids Res. 2018;46(D1):D239–45.
Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat Genet. 2007;39(10):1278–84.
Chen Y, Wang X. miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020;48(D1):D127–31.
Ziemann M, Kaspi A, El-Osta A. Evaluation of microRNA alignment techniques. RNA. 2016;22(8):1120–1138. https://doi.org/10.1261/rna.055509.115
Kamat MA, et al. PhenoScanner V2: an expanded tool for searching human genotype-phenotype associations, (in eng). Bioinformatics. 2019;35(22):4851–3. https://doi.org/10.1093/bioinformatics/btz469.
Sollis E, et al. The NHGRI-EBI GWAS Catalog: knowledge base and deposition resource. Nucleic Acids Res. 2023;51(D1):D977–85.
Relton CL, et al. Data resource profile: accessible resource for integrated epigenomic studies (ARIES). Int J Epidemiol. 2015;44(4):1181–90.
Bonder MJ, et al. Disease variants alter transcription factor levels and methylation of their binding sites. Nat Genet. 2017;49(1):131–8.
Martens JH, Stunnenberg HG. BLUEPRINT: mapping human blood cell epigenomes. Haematologica. 2013;98(10):1487.
Richardson TG, et al. Systematic Mendelian randomization framework elucidates hundreds of CpG sites which may mediate the influence of genetic variants on disease. Hum Mol Genet. 2018;27(18):3293–304.
Luijk R, et al. Autosomal genetic variation is associated with DNA methylation in regions variably escaping X-chromosome inactivation. Nat Commun. 2018;9(1):3738.
Porcu E, Rüeger S, Lepik K, Santoni FA, Reymond A, Kutalik Z. Mendelian randomization integrating GWAS and eQTL data reveals genetic determinants of complex and clinical traits. Nat Commun. 2019;10(1):3300.
Bahcall OG. GTEx pilot quantifies eQTL variation across tissues and individuals. Nat Rev Genet. 2015;16(7):375–375.
Yao C, et al. Genome-wide mapping of plasma protein QTLs identifies putatively causal genes and pathways for cardiovascular disease. Nat Commun. 2018;9(1):3268.
Lin E, Kuo PH, Liu YL, Yu YW, Yang AC, Tsai SJ. Prediction of antidepressant treatment response and remission using an ensemble machine learning framework. Pharmaceuticals (Basel). 2020;13(10):305. Published 2020 Oct 13. https://doi.org/10.3390/ph13100305
Vapnik V. The nature of statistical learning theory. Springer science & business media. 1999
Breiman L. Random forests. Mach Learn. 2001;45(1):5–32.
Loh WY. Classification and regression trees. Wiley Interdiscip Rev: Data Min Knowl Discov. 2011;1(1):14–23.
Le Cessie S, Van Houwelingen JC. Ridge estimators in logistic regression. Appl Stat. 1992;41(1):191–201.
Lundberg SM, Erion GG, Lee SI. Consistent individualized feature attribution for tree ensembles. arXiv preprint arXiv:1802.03888. 2018.
Zhou Y, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10(1):1523.
Wang DS, Dickson DW, Malter JS. beta-Amyloid degradation and Alzheimer's disease. J Biomed Biotechnol. 2006;2006(3):58406. https://doi.org/10.1155/JBB/2006/58406
Cardoso AL, et al. Towards frailty biomarkers: candidates from genes and pathways regulated in aging and age-related diseases, (in English). Ageing Res Rev. 2018;47:214–77. https://doi.org/10.1016/j.arr.2018.07.004.
Sahay A, Molliver ME, Ginty DD, Kolodkin AL. Semaphorin 3F is critical for development of limbic system circuitry and is required in neurons for selective CNS axon guidance events. J Neurosci. 2003;23(17):6671–80. https://doi.org/10.1523/JNEUROSCI.23-17-06671.2003.
Romera-Liebana L, Orfila F, Segura JM, et al. Effects of a primary care-based multifactorial intervention on physical and cognitive function in frail, elderly individuals: a randomized controlled trial. J Gerontol A Biol Sci Med Sci. 2018;73(12):1688–1674. https://doi.org/10.1093/gerona/glx259
Serra-Prat M, Sist X, Domenich R, et al. Effectiveness of an intervention to prevent frailty in pre-frail community-dwelling older people consulting in primary care: a randomised controlled trial. Age Ageing. 2017;46(3):401–407. https://doi.org/10.1093/ageing/afw242
Cameron ID, et al. A multifactorial interdisciplinary intervention reduces frailty in older people: randomized trial. BMC Med. 2013;11(1):1–10.
Eklund K, Wilhelmson K, Gustafsson H, Landahl S, Dahlin-Ivanoff S. One-year outcome of frailty indicators and activities of daily living following the randomised controlled trial;“Continuum of care for frail older people.” BMC Geriatr. 2013;13(1):1–10.
Blodgett J, Theou O, Kirkland S, Andreou P, Rockwood K. Frailty in NHANES: comparing the frailty index and phenotype. Arch Gerontol Geriatr. 2015;60(3):464–70.
Pulit SL, Stoneman C, Morris AP, et al. Meta-analysis of genome-wide association studies for body fat distribution in 694 649 individuals of European ancestry. Hum Mol Genet. 2019;28(1):166–174. https://doi.org/10.1093/hmg/ddy327
Cadby G, Giles C, Melton PE, et al. Comprehensive genetic analysis of the human lipidome identifies loci associated with lipid homeostasis with links to coronary artery disease. Nat Commun. 2022;13(1):3124. Published 2022 Jun 6. https://doi.org/10.1038/s41467-022-30875-7
Mirhafez SR, et al. Zinc Finger 259 gene polymorphism rs964184 is associated with serum triglyceride levels and metabolic syndrome, (in Eng). Int J Mol Cell Med. 2016;5(1):8–18.
Trinder M, Vikulova D, Pimstone S, Mancini GBJ, Brunham LR. Polygenic architecture and cardiovascular risk of familial combined hyperlipidemia. Atherosclerosis. 2022;340:35–43. https://doi.org/10.1016/j.atherosclerosis.2021.11.032
Wright KM, Rand KA, Kermany A, et al. A prospective analysis of genetic variants associated with human lifespan. G3 (Bethesda). 2019;9(9):2863–2878. Published 2019 Sep 4. https://doi.org/10.1534/g3.119.400448
Rajmohan V, Mohandas E. The limbic system, (in Eng). Indian J Psychiatry. 2007;49(2):132–9. https://doi.org/10.4103/0019-5545.33264.
Liu LK, et al. Cerebellar-limbic neurocircuit is the novel biosignature of physio-cognitive decline syndrome, (in Eng). Aging. 2020;12(24):25319–36. https://doi.org/10.18632/aging.104135.
Bulik-Sullivan BK, et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47(3):291–5.
Onopiuk A, Tokarzewicz A, Gorodkiewicz E. Cystatin C: a kidney function biomarker. Adv Clin Chem. 2015;68:57–69. https://doi.org/10.1016/bs.acc.2014.11.007
Veldurthy V, Wei R, Oz L, Dhawan P, Jeon YH, Christakos S. Vitamin D, calcium homeostasis and aging. Bone Res. 2016;4:16041. Published 2016 Oct 18. https://doi.org/10.1038/boneres.2016.41
McGill MR. The past and present of serum aminotransferases and the future of liver injury biomarkers. EXCLI J. 2016;15:817–828. Published 2016 Dec 15. https://doi.org/10.17179/excli2016-800
Kashani K, Rosner MH, Ostermann M. Creatinine: From physiology to clinical application. Eur J Intern Med. 2020;72:9–14. https://doi.org/10.1016/j.ejim.2019.10.025
Yin J, Tian L. Joint confidence region estimation for area under ROC curve and Youden index. Stat Med. 2014;33(6):985–1000.
Wang Q, Wang Y, Lehto K, Pedersen NL, Williams DM, Hägg S. Genetically-predicted life-long lowering of low-density lipoprotein cholesterol is associated with decreased frailty: a Mendelian randomization study in UK biobank. eBioMed. 2019;45:487–94. https://doi.org/10.1016/j.ebiom.2019.07.007.
Wong TY, Massa MS, O’Halloran AM, Kenny RA, Clarke R. Cardiovascular risk factors and frailty in a cross-sectional study of older people: implications for prevention. Age Ageing. 2018;47(5):714–20. https://doi.org/10.1093/ageing/afy080.
Mekli K, et al. Frailty Index associates with GRIN2B in two representative samples from the United States and the United Kingdom. PLOS One. 2018;13(11):e0207824. https://doi.org/10.1371/journal.pone.0207824.
Timmers PR, Mounier N, Lall K, et al. Genomics of 1 million parent lifespans implicates novel pathways and common diseases and distinguishes survival chances. Elife. 2019;8:e39856. Published 2019 Jan 15. https://doi.org/10.7554/eLife.39856
Pilling LC, Atkins JL, Bowman K, et al. Human longevity is influenced by many genetic variants: evidence from 75,000 UK Biobank participants. Aging (Albany NY). 2016;8(3):547–560. https://doi.org/10.18632/aging.100930
Davies G, et al. A genome-wide association study implicates the APOE locus in nonpathological cognitive ageing. Mol Psychiatry. 2014;19(1):76–87.
McKay GJ, et al. Variations in apolipoprotein E frequency with age in a pooled analysis of a large group of older people. Am J Epidemiol. 2011;173(12):1357–64.
Gerdes LU, Jeune B, Ranberg KA, Nybo H, Vaupel JW. Estimation of apolipoprotein E genotype-specific relative mortality risks from the distribution of genotypes in centenarians and middle-aged men: apolipoprotein E gene is a "frailty gene," not a "longevity gene". Genet Epidemiol. 2000;19(3):202–210. https://doi.org/10.1002/1098-2272(200010)19:33.0.CO;2-Q
Mourtzi N, Ntanasi E, Yannakoulia M, et al. Apolipoprotein ε4 allele is associated with frailty syndrome: results from the hellenic longitudinal investigation of ageing and diet study. Age Ageing. 2019;48(6):917–921. https://doi.org/10.1093/ageing/afz098
Sathyan S, Verghese J. Genetics of frailty: a longevity perspective. Transl Res. 2020;221:83–96. https://doi.org/10.1016/j.trsl.2020.03.005.
Chhetri J, et al. Apolipoprotein E polymorphism and frailty: apolipoprotein ε4 allele is associated with fatigue but not frailty syndrome in a community-dwelling older population cohort. J Nutr Health Aging. 2021;25:410–5.
Garatachea N, et al. ApoE gene and exceptional longevity: insights from three independent cohorts, (in Eng). Exp Gerontol. 2014;53:16–23. https://doi.org/10.1016/j.exger.2014.02.004.
Ryu S, Atzmon G, Barzilai N, Raghavachari N, Suh Y. Genetic landscape of APOE in human longevity revealed by high-throughput sequencing, (in eng). Mech Ageing Dev. 2016;155:7–9. https://doi.org/10.1016/j.mad.2016.02.010.
Safieh M, Korczyn AD, Michaelson DM. ApoE4: an emerging therapeutic target for Alzheimer's disease. BMC Med. 2019;17(1):64. Published 2019 Mar 20. https://doi.org/10.1186/s12916-019-1299-4
McKay GJ, Silvestri G, Chakravarthy U, Dasari S, Fritsche LG, Weber BH, Keilhauer CN, Klein ML, Francis PJ, Klaver CC, Vingerling JR, Ho L, De Jong PT, Dean M, Sawitzke J, Baird PN, Guymer RH, Stambolian D, Orlin A, Seddon JM, Patterson CC. Variations in apolipoprotein E frequency with age in a pooled analysis of a large group of older people. American J Epidemiol. 2011;173(12):1357–1364. https://doi.org/10.1093/aje/kwr015
Fry A, Littlejohns TJ, Sudlow C, et al. Comparison of sociodemographic and health-related characteristics of uk biobank participants with those of the general population. Am J Epidemiol. 2017;186(9):1026–1034. https://doi.org/10.1093/aje/kwx246
Acknowledgements
This research has been conducted using data from UK Biobank (www.ukbiobank.ac.uk), a major biomedical database, approved under project # 54423.
Funding
This work was supported by China Medical University Hospital, Taichung, Taiwan (C1100708016) and National Science and Technology Council, Taiwan (NSTC 111-2634-F-002-017). The funders had no role in the design and conduct of the study; collection, management, analysis and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Watson Hua-Sheng Tseng and Amrita Chattopadhyay are co-first authors with equal contribution.
Supplementary Information
Below is the link to the electronic supplementary material.
About this article
Cite this article
Tseng, W.HS., Chattopadhyay, A., Phan, N.N. et al. Utilizing multimodal approach to identify candidate pathways and biomarkers and predicting frailty syndrome in individuals from UK Biobank. GeroScience 46, 1211–1228 (2024). https://doi.org/10.1007/s11357-023-00874-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11357-023-00874-7