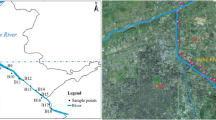
Abstract
Water samples for the 16S rRNA gene and water quality analyses were collected from around 155 km of river segments surrounding the urban areas in Xi’an, China. Multiple statistical analyses showed that the dynamic shifts of microbial communities in the Chan, Ba, and Feng Rivers from the spring to the summer seasons were apparent but little in the Zao River. The heterogeneity of microbial distributions was more due to the influence of hydrologic conditions and various sources of inflows in the rivers. The LEfSe analysis showed that the Chan and Zao Rivers, both more impacted by the sewage effluents, were more differentially abundant with bacteria related to polluted water, but the Ba and Feng Rivers, both on the outer side of the city, were more abundant with microbial communities in soil and freshwater environments in the summer. Multiple statistical analyses indicated that environmental variables had significant impacts on microbial communities. The geographical information system-based spatial analysis showed heterogeneity of microbial community distributions along the rivers. This study showed that the high-throughput sequencing analysis could identify some pathogenic bacteria that would significantly threaten public health and eco-environments in urban rivers.








Similar content being viewed by others
Data availability
The datasets generated and analyzed during the current study are submitted with this article.
References
Araya R, Tani K, Takagi T, Yamaguchi N, Nasu M (2003) Bacterial activity and community composition in stream water and biofilm from an urban river determined by fluorescent in situ hybridization and DGGE analysis. FEMS Microbiol Ecol 43(1):111–119
Bai Y, Wang Q, Liao K, Jian Z, Zhao C, Qu J (2018) Fungal community as a bioindicator to reflect anthropogenic activities in a river ecosystem. Front Microbiol 9:3152
Berglund B, Fick J, Lindgren PE (2015) Urban wastewater effluent increases antibiotic resistance gene concentrations in a receiving northern European river. Environ Toxicol Chem 34(1):192–196
Bower PA, Scopel CO, Jensen ET, Depas MM, McLellan SL (2005) Detection of genetic markers of fecal indicator bacteria in Lake Michigan and determination of their relationship to Escherichia coli densities using standard microbiological methods. Appl Environ Microbiol 71(12):8305–8313
Caporaso JG, J Kuczynski et al (2010) Nature Methods https://doi.org/10.1038/nmeth.f.303
CDC (2020) Sexually Transmitted Disease Surveillance. Atlanta, GA: Department of Health and Human Services; April 2022
Chaudhary A et al (2018) "Taxon-driven functional shifts associated with storm flow in an urban stream microbial community." mSphere 3(4)
Daims H, Lebedeva E, Pjevac P et al (2015) Complete nitrification by Nitrospira bacteria. Nature 528:504–509. https://doi.org/10.1038/nature16461
Drury B, Rosi-Marshall E, Kelly JJ (2013) Wastewater treatment effluent reduces the abundance and diversity of benthic bacterial communities in urban and suburban rivers. Appl Environ Microbiol 79(6):1897–1905
Edgar RC (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26(19):2460–2461
Edgar RC (2013) UPARSE: Highly accurate OTU sequences from microbial amplicon reads, Nature Methods 10.1038/ nmeth.2604
Eraqi WA et al (2018) The Nile River microbiome reveals a remarkably stable community between wet and dry seasons, and sampling sites, in a large urban metropolis (Cairo, Egypt). Omics J Integr Biol 22(8):553–564
Gauch HG Jr (1982) Multivariate analysis in community ecology. Cambridge University Press, Cambridge, p 298
Guan Y, Jia J, Wu L, Xue X, Zhang G, Wang Z (2018) Analysis of bacterial community characteristics, abundance of antibiotics and antibiotic resistance genes along a pollution gradient of Ba river in Xi’an. China Front Microbiol 9:3191
Matheus Carnevali PB, Schulz F, Castelle CJ et al (2019) Hydrogen-based metabolism as an ancestral trait in lineages sibling to the Cyanobacteria. Nat Commun 10:463. https://doi.org/10.1038/s41467-018-08246-y
Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ (2015) IQ-TREE: a fast and effective stochastic algorithm for estimating maximum likelihood phylogenies. Mol Biol Evol 32:268–274
Peng FJ, Pan CG, Zhang NS, Ter Braak CJF, Salvito D, Selck H, Ying GG, Van den Brink PJ (2019) Benthic invertebrate and microbial biodiversity in sub-tropical urban rivers: correlations with environmental variables and emerging chemicals. Sci Total Environ 709:136281
Schloss PD (2020) Reintroducing mothur: 10 years later. Appl Environ Microbiol 86:e02343-e2419. https://doi.org/10.1128/AEM.02343-19
Segata N, Izard J, Waldron L, et al (2011) Metagenomic biomarker discovery and explanation. Genome Biol 12(6):R60. Published 2011 Jun 24 https://doi.org/10.1186/gb-2011-12-6-r60.
Whittaker RH (1960) Vegetation of the Siskiyou mountains, Oregon and California. Ecol Monogr 30:279–338. https://doi.org/10.2307/1943563
Whittaker RH (1972) Evolution and measurement of species diversity. Taxon 21:213–251. https://doi.org/10.2307/1218190
Youssef NH, Rinke C, Stepanauskas R, Farag I, Woyke T, Elshahed MS (2015) Insights into the metabolism, lifestyle and putative evolutionary history of the novel archaeal phylum ‘Diapherotrites.’ ISME J 9(2):447–460. https://doi.org/10.1038/ismej.2014.141
Zhang J, Liu S, Cuiju L, Luan J, Liu X, Xinwei G, Niu B (2021) Soil bacterial and fungal richness and network exhibit different responses to long-term throughfall reduction in a warm-temperate oak forest. Forests 12:165. https://doi.org/10.3390/f12020165
Zhang H, Wang Y, Chen S, Zhao Z, Feng J, Zhang Z, Lu K, Jia J (2018) Water bacterial and fungal community compositions associated with urban lakes, Xi’an, China. Int J Environ Res Public Health 15(3)
Zhou J et al (2013) Random sampling process leads to an overestimation of beta-diversity of microbial communities. Mbio 4(3):e00324-e313
Acknowledgements
Great thanks to the editors and anonymous reviewers for their help in improving this paper.
Funding
Funding for this research was provided by the National Key Research and Development Programs of China (Grant No. 2021YFC3201100), the Shaanxi Key R&D Plan International Science and Technology Cooperation and Exchange Program (Grant No. 2020KWZ-023), and the “Light of West China” Program of the Chinese Academy of Sciences.
Author information
Authors and Affiliations
Contributions
YL directed the research and wrote the manuscript, and YL and LZ conducted data analysis and prepared the tables and figures. LZ, HL, YF, RL, XC, YL, and XL participated in water sample collections and analyses. All authors read the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Responsible Editor: Robert Duran
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yanqing Lian and Lisha Zhen are the co-first author.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Lian, Y., Zhen, L., Chen, X. et al. Microbial biomarkers as indication of dynamic and heterogeneous urban water environments. Environ Sci Pollut Res 30, 107304–107316 (2023). https://doi.org/10.1007/s11356-022-24539-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-022-24539-8