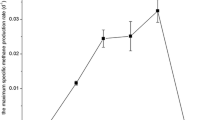
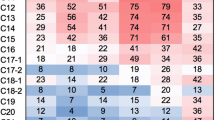
Abstract
Microbial community composition and metabolic potential have been explored in petroleum-hydrocarbon-contaminated sludge of an oil storage facility. Culture-independent clone library-based 16S rRNA gene analyses revealed that the bacterial community within the sludge was dominated by the members of β-Proteobacteria (35 %), followed by Firmicutes (13 %), δ-Proteobacteria (11 %), Bacteroidetes (10 %), Acidobacteria (6 %), α-Proteobacteria (3 %), Lentisphaerae (2 %), Spirochaetes (2 %), and unclassified bacteria (5 %), whereas the archaeal community was composed of Thermoprotei (54 %), Methanocellales (33 %), Methanosarcinales/Methanosaeta (8 %) and Methanoculleus (1 %) members. Methyl coenzyme M reductase A (mcrA) gene (a functional biomarker) analyses also revealed predominance of hydrogenotrophic, methanogenic Archaea (Methanocellales, Methanobacteriales and Methanoculleus members) over acetoclastic methanogens (Methanosarcinales members). In order to explore the cultivable bacterial population, a total of 28 resident strains were identified and characterized in terms of their physiological and metabolic capabilities. Most of these could be taxonomically affiliated to the members of the genera Bacillus, Paenibacillus, Micrococcus, Brachybacterium, Aerococcus, and Zimmermannella, while two strains were identified as Pseudomonas and Pseudoxanthomonas. Metabolic profiling exhibited that majority of these isolates were capable of growing in presence of a variety of petroleum hydrocarbons as sole source of carbon, tolerating different heavy metals at higher concentrations (≥1 mM) and producing biosurfactant during growth. Many strains could grow under a wide range of pH, temperature, or salinity as well as under anaerobic conditions in the presence of different electron acceptors and donors in the growth medium. Correlation between the isolates and their metabolic properties was estimated by the unweighted pair group method with arithmetic mean (UPGMA) analysis. Overall observation indicated the presence of diverse groups of microorganisms including hydrocarbonoclastic, nitrate reducing, sulphate reducing, fermentative, syntrophic, methanogenic and methane-oxidizing bacteria and Archaea within the sludge community, which can be exploited for in situ bioremediation of the oily sludge.







Similar content being viewed by others
References
Allen JP, Atekwana EA, Atekwana EA, Duris JW, Rossbach S (2007) The microbial community structure in petroleum-contaminated sediments corresponds to geophysical signatures. Appl Environ Microbiol 73:2860–2870
Almeida CMR, Reis I, Couto MN, Bordalo AA, Mucha AP (2013) Potential of the microbial community present in an unimpacted beach sediment to remediate petroleum hydrocarbons. Environ Sci Pollut Res 20:3176–3184
Banerjee A, Ghoshal AK (2010) Isolation and characterization of hyper tolerant Bacillus sp. from oil refinery and exploration sites. J Hazard Mater 176:85–91
Biddle JF, Lipp JS, Lever MA, Lloyd KG, Sørensen KB, Anderson R, Fredricks LF, Elvert M, Kelly TJ, Schrag DP, Sogin ML, Brenchley JE, Teske A, House CH, Hinrichs K-U (2006) Heterotrophic Archaea dominate sedimentary subsurface ecosystems off Peru. Proc Natl Acad Sci U S A 103:3846–3851
Borrell G, Lehours AC, Crouzet O, Jezequel D, Rockne K, Kulczak A, Duffaud E, Joblin K, Fonty G (2012) Stratification of Archaea in the deep sediments of a freshwater meromictic lake: vertical shift from methanogenic to uncultured archaeal lineages. PLoS ONE 7:433–446
Bugg T, Foght JM, Pickard AM, Gray RM (2000) Uptake and active efflux of polycyclic aromatic hydrocarbons by Pseudomonas fluorescens LP6a. Appl Environ Microbiol 66:5387–5392
Buriánková I, Brablcová L, Mach V, Dvořák P, Chaudhary PP, Rulík M (2013) Identification of Methanogenic archaea in the hyporheic sediment of Sitka stream. PLoS ONE 8:e80804
Cerqueira VS, Hollenbach EB, Maboni F, Vainstein MH, Camargo FAO, Peralbo MCR, Bento FM (2011) Biodegradation potential of oily sludge by pure and mixed bacterial cultures. Bioresour Technol 102:11003–11010
Choi EJ, Jin HM, Lee SH, Math RK, Madsen EL, Jeon CO (2013) Comparative genomic analysis and benzene, toluene, ethylbenzene, and o-, m-, and p-xylene (BTEX) degradation pathways of Pseudoxanthomonas spadix BD-a59. Appl Environ Microbiol 79:663–671
Chouari R, Paslier DL, Daegelen P, Dauga C, Weissenbach J, Sghir A (2010) Molecular analyses of the microbial community composition of an anoxic basin of a municipal wastewater treatment plant reveal a novel lineage of Proteobacteria. Microb Ecol 60:272–281
Daane LL, Harjono I, Zylstra JG, Haggblom MM (2001) Isolation and characterization of polycyclic aromatic hydrocarbon-degrading bacteria associated with the rhizosphere of salt marsh plants. Appl Environ Microbiol 67:2683–2691
Daffonchio D, Ferrer M, Mapelli F, Cherif A, Lafraya A, Malkawi HI, Yakimov MM, Abdel-Fattah YR, Blaghen M, Golyshin PN, Kalogerakis N, Boon N, Magagnini M, Fava F (2013) Bioremediation of Southern Mediterranean oil polluted sites comes of age. New Biotechnol 30:743–748
de Vasconcellos SP, Crespim E, da GF C, Senatore DB, Simioni KCM, Neto EVdos S, Marsaioli AJ, de VM O (2009) Isolation, biodegradation ability and molecular detection of hydrocarbon degrading bacteria in petroleum samples from a Brazilian offshore basin. Org Geochem 40:574–588
DeLong EF (1992) Archaea in coastal marine environments. Proc Nalt Acad Sci USA 89:5685–5689
Dhillon A, Lever M, Lloyd KG, Albert DB, Sogin ML, Teske A (2005) Methanogen diversity evidenced by molecular characterization of methyl coenzyme M reductase A (mcrA) genes in hydrothermal sediments of the Guaymas Basin. Appl Environ Microbiol 71:4592–4601
Ding B, Schmeling S, Fuchs G (2008) Anaerobic metabolism of catechol by the denitrifying bacterium Thauera aromatic—a result of promiscuous enzymes and regulators? J Bacteriol 190:1620–1630
Ferrera-Rodríguez O, Greer CW, Juck D, Consaul LL, Martıínez-Romero E, Whyte LG (2013) Hydrocarbon-degrading potential of microbial communities from Arctic plants. J Appl Microbiol 114:71–83
Gan Y, Qiu Q, Liu P, Lu Y (2012) Syntrophic oxidation of propionate in rice field soil at 15 and 30 °C under methanogenic conditions. Appl Environ Microbiol 78:4923–4932
Garcia JL, Patel BKC, Ollivier B (2000) Taxonomic phylogenetic and ecological diversity of methanogenic Archaea. Anaerobe 6:205–226
George IF, Liles MR, Hartmann M, Ludwig W, Goodman RM, Agathos NS (2009) Changes in soil Acidobacteria communities after 2,4,6-trinitrotoluene contamination. FEMS Microbiol Lett 296:159–166
Gerdes B, Brinkmeyer R, Dieckmann G, Helmke E (2005) Influence of crude oil on changes of bacterial communities in Artic sea-ice. FEMS Microbiol Ecol 53:129–139
Gillespie IMM, Philp JC (2013) Bioremediation, an environmental remediation technology for the bioeconomy. Trends Biotechnol 31:329–332
Good I (1953) The population frequencies of species and the estimation of population parameters. Biometrika 40:237–264
Grabowski A, Nercession O, Fayolle F, Blanchet D, Jeanthon C (2005) Microbial diversity in production water of a low-temperature biodegraded oil reservoir. FEMS Microbiol Ecol 54:427–443
Gray ND, Sherry A, Grant AK, Rowan AK, Hubert CRJ, Callbeck CM, Aitken CM, Jones DM, Adams JJ, Larter SR, Head IM (2011) The quantitative significance of Syntrophaceae and syntrophic partnerships in methanogenic degradation of crude oil alkanes. Environ Microbiol 13:2957–2975
Hales BA, Edwards C, Ritchie DA, Hall G, Pickup RW, Saunders JR (1996) Isolation and identification of methanogen-specific DNA from blanket bog peat by PCR amplification and sequence analysis. Appl Environ Microbiol 62:668–675
Hazen TC, Rocha AM, Techtmann SM (2013) Advances in monitoring environmental microbes. Curr Opin Biotechnol 24:526–533
Hernandez-Raquet G, Budzinski, Caumette P, Dabert P, Ménach KL, Muyzer G, Duran R (2006) Molecular diversity studies of bacterial communities of oil polluted microbial mats from the Etang de Berre (France). FEMS Microbiol Ecol 58:550–562
Im WT, Bae HS, Yokota A, Lee ST (2004) Herbaspirillum chlorophenolicum sp. nov., a 4-chlorophenol-degrading bacterium. Int J Syst Evol Microbiol 54:851–855
Imfeld G, Aragonés CE, Fetzer I, Mészáros E, Zeiger S, Nijenhuis I, Nikolausz M, Delercel S, Richnow HH (2010) Characterization of microbial communities in the aqueous phase of a constructed model wetland treating 1,2-dichloroethene-contaminated groundwater. FEMS Microbiol Ecol 72:74–88
Inoue K, Habe H, Yamane H, Omori T, Nojiri H (2005) Diversity of carbazole-degrading bacteria having the car gene cluster: isolation of a novel gram-positive carbazole-degrading bacterium. FEMS Microbiol Lett 245:145–153
Jeanthon C, L’Haridon S, Cueff V, Banta A, Reysenbach AL, Prieur D (2002) Thermodesulfobacterium hydrogeniphilum sp. nov., a thermophilic, chemolithoautotrophic, sulfate-reducing bacterium isolated from a deep-sea hydrothermal vent at Guaymas Basin, and emendation of the genus Thermodesulfobacterium. Int J Syst Evol Microbiol 52:765–772
Jukes TH, Cantor CR (1969) Evolution of protein molecules. In: Munro HN (ed) Mammalian protein metabolism. Academic Press, New York, pp 21–132
Kasai Y, Takahata Y, Watanabe K (2005) Physiological and molecular characterization of a microbial community established in unsaturated, petroleum-contaminated soil. Environ Microbiol 7:806–818
Kazy SK, Sar P, Asthana RK, Singh SP (1999) Copper uptake and its compartmentalization in Pseudomonas aeruginosa strains: chemical nature of cellular metal. W J Microbiol Biotechnol 15:599–605
Kim JM, Le NT, Chung BS, Park JH, Jin-W B, Madsen EL, Jeon CO (2008) Influence of soil components on the biodegradation of benzene, toluene, ethylbenzene, and o-, m-, and p-xylenes by the newly isolated bacterium Pseudoxanthomonas spadix BD-a59. Appl Environ Microbiol 74:7313–7320
Kleikemper J, Pombo SA, Schroth MH, Sigler WV, Pesaro M, Zeyer J (2005) Activity and diversity of methanogens in a petroleum hydrocarbon-contaminated aquifer. Appl Environ Microbiol 71:149–158
Kleinsteuber S, Schleinitz KM, Breitfeld J, Harms H, Richnow HH, Vogt C (2008) Molecular characterization of bacterial communities mineralizing benzene under sulfate-reducing conditions. FEMS Microbiol Ecol 66:143–157
Kleinsteuber S, Schleinitz KM, Vogt C (2012) Key players and team play: anaerobic microbial communities in hydrocarbon-contaminated aquifers. Appl Microbiol Biotechnol 94:851–873
Kobayashi H, Endo K, Sakata S, Mayumi D, Kawaguchi H, Ikarashi M, Miyagawa Y, Maeda H, Sato K (2012) Phylogenetic diversity of microbial communities associated with the crude-oil, large-insoluble-particle and formation-water components of the reservoir fluid from a non-flooded high-temperature petroleum reservoir. J Biosci Bioeng 113:204–210
Kostka JE, Prakash O, Overholt WA, Green SJ, Freyer G, Canion A, Delgardio J, Norton N, Hazen TC, Huettel M (2011) Hydrocarbon-degrading bacteria and the bacterial community response in Gulf of Mexico beach sands impacted by the Deepwater Horizon oil spill. Appl Environ Microbiol 77:7962–7974
Kunapuli U, Lueders T, Meckenstock RU (2007) The use of stable isotope probing to identify key iron-reducing microorganisms involved in anaerobic benzene degradation. ISME J 1:643–653
LaMontagne MG, Leifer L, Bergman S, Van De Werfhorst LC, Holden PA (2004) Bacterial diversity in marine hydrocarbon seep sediments. Environ Microbiol 6:799–808
Lane DJ (1991) 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M (eds) Nucleic acid techniques in bacterial systematics. Wiley, New York, pp 115–175
Lee SH, Jin HM, Lee HJ, Kim JM, Jeon CO (2012) Complete genome sequence of the BTEX-degrading bacterium Pseudoxanthomonas spadix BD-a59. J Bacteriol 194:544. doi:10.1128/JB.06436-11
Li Q, Wang F, Chen Z, Yin X, Xiao X (2012) Stratified active archaeal communities in the sediments of Jiulong River estuary, China. Front Microbiol 3:311. doi:10.3389/fmicb.2012.00311
Liu R, Zhang Y, Ding R, Li D, Goa Y, Yang M (2009) Comparison of archaeal and bacterial community structures in heavily oil-contaminated and pristine soils. J Biosci Bioeng 108:400–407
Liu J, Wu W, Chen C, Sun F, Chen Y (2011) Prokaryotic diversity, composition structure, and phylogenetic analysis of microbial communities in leachate sediment ecosystems. Appl Microbiol Biotechnol 91:1659–1675
Lliro´s M, Casamayor EO, Borrego C (2008) High archaeal richness in the water column of a freshwater sulfurous karstic lake along an interannual study. FEMS Microbiol Ecol 66:331–342
Lors C, Ryngaert A, Perie F, Diels L, Damidot D (2010) Evolution of bacterial community during bioremediation of PAHs in a coal tar contaminated soil. Chemosphere 81:1263–1271
Luton PE, Wayne JM, Sharp RJ, Riley PW (2002) The mcrA gene as an alternative to 16S rRNA in the phylogenetic analysis of methanogen populations in landfill. Microbiol 148:3521–3530
Mao Y, Zhang X, Xia X, Zhong H, Zhao L (2010) Versatile aromatic compound-degrading capacity and microdiversity of Thauera strains isolated from a coking wastewater treatment bioreactor. J Ind Microbiol Biotechnol 37:927–934
Mayumi D, Mochimaru H, Yoshioka H, Sakata S, Maeda H, Miyagawa Y, Ikarashi M, Takeuchi M, Kamagata Y (2011) Evidence for syntrophic acetate oxidation coupled to hydrogenotrophic methanogenesis in the high-temperature petroleum reservoir of Yabase oil field (Japan). Environ Microbiol 13:1995–2006
Melcher RJ, Apitz SE, Hemmingsen BB (2002) Impact of irradiation and polycyclic aromatic hydrocarbon spiking on microbial populations in marine sediment for future aging and biodegradability studies. Appl Environ Microbiol 68:2858–2868
Militon C, Boucher D, Vachelard C, Perchet G, Barra V, Troquet J, Peyretaillade E, Peyret P (2010) Bacterial community changes during bioremediation of aliphatic hydrocarbon-contaminated soil. FEMS Microbiol Ecol 74:669–681
Mishra S, Jyoti J, Kuhad RC, Lal B (2001) In situ bioremediation potential of an oily sludge-degrading bacterial consortium. Curr Microbiol 43:328–335
Mukherjee AK, Bordoloi NK (2012) Biodegradation of benzene, toluene, and xylene (BTX) in liquid culture and in soil by Bacillus subtilis and Pseudomonas aeruginosa strains and a formulated bacterial consortium. Environ Sci Pollut Res 19:3380–3388
Mulla SI, Hoskeri RS, Shouche YS, Ninnekar HZ (2011) Biodegradation of 2-nitrotoluene by Micrococcus sp. strain SMN-1. Biodegradation 22:95–102
Müller D, Simeonova DD, Riegel P, Mangenot S, Koechler S, Lievremont D, Bertin PN, Lett MC (2006) Herminiimonas arsenicoxydans sp. nov., a metalloresistant bacterium. Int J Syst Evol Microbiol 56:1765–1769
Najafi AR, Rahimpour MR, Jahanmiri AH, Roostaazad R, Arabian D (2011) Interactive optimization of biosurfactant production by Paenibacillus alvei ARN63 isolated from an Iran oil well. Colloids Surf B: Biointerfaces 82:33–39
Narancic T, Kenny ST, Djokic L, Vasiljevic B, O’Connor KE, Nikodinovic-Runic J (2012) Medium-chain-length polyhydroxyalkanoate production by newly isolated Pseudomonas sp. TN301 from a wide range of polyaromatic and monoaromatic hydrocarbons. J Appl Microbiol 113:508–520
Patel V, Cheturvedula S, Madamwar D (2012) Phenanthrene degradation by Pseudoxanthomonas sp. DMVP2 isolated from hydrocarbon contaminated sediment of Amlakhadi canal, Gujarat, India. J Hazard Mater 201–202:43–51
Penner TJ, Foght JM (2010) Mature fine tailings from oil sands processing harbour diverse methanogenic communities. Can J Microbiol 56:459–70
Pham VD, Hnatow LL, Zhang S, Fallon RD, Jackson SC (2009) Characterizing microbial diversity in production water from an Alaskan mesothermic petroleum reservoir with two independent molecular methods. Environ Microbiol 11:176–187
Popp N, Schlomann M, Mau M (2006) Bacterial diversity in the active stage of a bioremediation system for mineral oil hydrocarbon-contaminated soils. Microbiology 152:3291–3304
Reddy MV, Devi MP, Chandrasekhar K, Goud RK, Mohan SV (2011) Aerobic remediation of petroleum sludge through soil supplementation: microbial community analysis. J Hazard Mater 197:80–87
Ren HY, Zhang XJ, Z-yong S, Rupert W, Gao GJ (2011) Comparison of microbial community compositions of injection and production well samples in a long-term water-flooded petroleum reservoir. PLoS ONE 6:e23258
Rouviere PE, Chen MW (2003) Isolation of Brachymonas petroleovorans CHX, a novel cyclohexane-degrading beta-proteobacterium. FEMS Microbiol Lett 227:101–106
Sakai M, Ezaki S, Suzuki N, Kurane R (2005) Isolation and characterization of a novel polychlorinated biphenyl-degrading bacterium, Paenibacillus sp. KBC101. Appl Microbiol Biotechnol 68:111–116
Sakai S, Imachi H, Hanada S, Ohashi A, Harada H, Kamagata Y (2008) Methanocella paludicola gen. nov., sp. nov., a methane-producing archaeon, the first isolate of the lineage ‘Rice Cluster I’, and proposal of the new archaeal order Methanocellales ord. nov. Int J Syst Evol Microbiol 58:929–936
Sambrook J, Fritsch EF, Maniatis T (1990) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory Press, New York, p 73
Silva TR, Verde LCL, Santos Neto EV, Oliveira VM (2013) Diversity analyses of microbial communities in petroleum samples from Brazilian oil fields. Int Biodeterior Biodegrad 81:57–70
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Bio Evol 24:1596–1599
van der Kraan GM, Bruining J, Lomans BP, van Loosdrecht MCM, Muyzer G (2010) Microbial diversity of an oil-water processing site and its associated oil field: the possible role of microorganisms as information carriers from oil-associated environments. FEMS Microbiol Ecol 71:428–443
van der Zaan BM, Saia FT, Stams AJM, Plugge CM, de Vos WM, Smidt H, Langenhoff AAM, Gerritse J (2012) Anaerobic benzene degradation under denitrifying conditions: Peptococcaceae as dominant benzene degraders and evidence for a syntrophic process. Environ Microbiol 14:1171–1181
Wang L, Wang W, Lai Q, Shao Z (2010) Gene diversity of CYP153A and AlkB alkane hydroxylases in oil-degrading bacteria isolated from the Atlantic Ocean. Environ Microbiol 12:1230–1242
Wolicka D, Borkowski A, Dobrzyński D (2010) Interactions between microorganisms, crude oil and formation waters. Geomicrobiol J 27:430–452
Yan Z, Song N, Cai H, Tay JH, Jiang H (2012) Enhanced degradation of phenanthrene and pyrene in freshwater sediments by combined employment of sediment microbial fuel cell and amorphous ferric hydroxide. J Hazard Mater 199–200:217–225
Zhang DC, Mörtelmaier C, Margesin R (2012) Characterization of the bacterial archaeal diversity in hydrocarbon-contaminated soil. Sci Total Environ 421–422:184–196
Acknowledgments
This work was financially supported by the Department of Science and Technology, Government of India under Fast Track Project for Young Scientist scheme (SR/FT//LS-078/2008). The authors gratefully acknowledge the support for obtaining oily sludge sample from Bharat Petroleum Corporation Limited, Rajbandh (TOP), Durgapur, West Bengal, India. The authors are thankful to Dr. Pinaki Sar, Department of Biotechnology, Indian Institute of Technology Kharagpur, for his critical comments on the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible editor: Gerald Thouand
Rights and permissions
About this article
Cite this article
Das, R., Kazy, S.K. Microbial diversity, community composition and metabolic potential in hydrocarbon contaminated oily sludge: prospects for in situ bioremediation. Environ Sci Pollut Res 21, 7369–7389 (2014). https://doi.org/10.1007/s11356-014-2640-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-014-2640-2