Abstract
Purpose
Macropinocytosis serves as a highly conserved endocytotic process that has recently been shown as a critical mechanism by which RAS-transformed cells transport extracellular protein into intracellular amino acid pathways to support their unique metabolic needs. We developed NIR fluorescently labeled molecular imaging probes to monitor macropinocytosis-mediated uptake of albumin in a K-RAS-dependent manner.
Procedures
Using western blot analysis, immunofluorescence, and flow cytometry, albumin retention was characterized in vitro across several RAS-activated lung and pancreatic cancer cell lines. AF790-albumin was synthesized and administered to mice bearing K-RAS mutant xenograft tumors of H460 (K-RAS p.Q61H) and H358 (K-RAS p.G12C) non-small cell lung cancers on each flank. Mice were treated daily with 2 mg/kg of ARS-1620, a targeted RAS p.G12C inhibitor, for 2 days and imaged following each treatment. Subsequently, the mice were then treated daily with 10 mg/kg of amiloride, a general inhibitor of macropinocytosis, for 2 days and imaged. Intratumoral distribution of AF790-albumin was assessed in vivo using near-infrared (NIR) fluorescence imaging.
Results
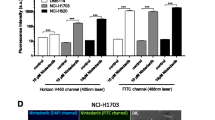
Albumin retention was observed as a function of K-RAS activity and macropinocytosis across several lung and pancreatic cancer cell lines. We documented that ARS-1620-induced inhibition of K-RAS activity or amiloride-mediated inhibition of macropinocytosis significantly reduced albumin uptake. Tumor retention in vivo of AF790-albumin was both RAS inhibition-dependent as well as abrogated by inhibition of macropinocytosis.
Conclusions
These data provide a novel approach using NIR-labeled human serum albumin to identify and monitor RAS-driven tumors as well as evaluate the on-target efficacy in vivo of inhibitors, such as ARS-1620.






Similar content being viewed by others
References
Prior IA, Hood FE, Hartley JL (2020) The frequency of Ras mutations in cancer. Cancer Res 80(14):2969–2974. https://doi.org/10.1158/0008-5472.CAN-19-3682
Cox AD et al (2014) Drugging the undruggable RAS: mission possible? Nat Rev Drug Discov 13(11):828–851
Downward J (2003) Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer 3(1):11–22
Lim JKM, Leprivier G (2019) The impact of oncogenic RAS on redox balance and implications for cancer development. Cell Death Dis 10(12):955
Simanshu DK, Nissley DV, McCormick F (2017) RAS proteins and their regulators in human disease. Cell 170(1):17–33
Kitajima S, Thummalapalli R, Barbie DA (2016) Inflammation as a driver and vulnerability of KRAS mediated oncogenesis. Semin Cell Dev Biol 58:127–135
Janes MR et al (2018) Targeting KRAS Mutant Cancers with a Covalent G12C-Specific Inhibitor. Cell 172(3):578-589 e17
Canon J et al (2019) The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature 575(7781):217–223
Hallin J et al (2020) The KRAS(G12C) Inhibitor MRTX849 provides insight toward therapeutic susceptibility of KRAS-mutant cancers in mouse models and patients. Cancer Discov 10(1):54–71
Wu X et al (2013) Inhibition of RAS-effector interaction by cyclic peptides. Medchemcomm 4(2):378–382
Shima F et al (2013) In silico discovery of small-molecule RAS inhibitors that display antitumor activity by blocking the RAS-effector interaction. Proc Natl Acad Sci U S A 110(20):8182–8187
Kauke MJ et al (2017) An engineered protein antagonist of K-RAS/B-RAF interaction. Sci Rep 7(1):5831
Spencer-Smith R et al (2017) Inhibition of RAS function through targeting an allosteric regulatory site. Nat Chem Biol 13(1):62–68
Spencer-Smith R, O’Bryan JP (2019) Direct inhibition of RAS: quest for the holy grail? Semin Cancer Biol 54:138–148
Sutton MN et al (2019) DIRAS3 (ARHI) Blocks RAS/MAPK signaling by binding directly to RAS and disrupting RAS clusters. Cell Rep 29(11):3448-3459 e6
Yoshida S et al (2018) Macropinocytosis, mTORC1 and cellular growth control. Cell Mol Life Sci 75(7):1227–1239
Feitelson MA et al (2015) Sustained proliferation in cancer: mechanisms and novel therapeutic targets. Semin Cancer Biol 35(Suppl):S25–S54
Commisso C, Flinn RJ, Bar-Sagi D (2014) Determining the macropinocytic index of cells through a quantitative image-based assay. Nat Protoc 9(1):182–192
Commisso C et al (2013) Macropinocytosis of protein is an amino acid supply route in RAS-transformed cells. Nature 497(7451):633–637
Swiercz R et al (2017) Loss of expression of the recycling receptor, FcRn, promotes tumor cell growth by increasing albumin consumption. Oncotarget 8(2):3528–3541
Nakase I et al (2015) Active macropinocytosis induction by stimulation of epidermal growth factor receptor and oncogenic RAS expression potentiates cellular uptake efficacy of exosomes. Sci Rep 5:10300
Seton-Rogers S (2020) KRAS-G12C in the crosshairs. Nat Rev Cancer 20(1):3
Nguyen DX, Bos PD, Massague J (2009) Metastasis: from dissemination to organ-specific colonization. Nat Rev Cancer 9(4):274–284
Keenan MM, Chi JT (2015) Alternative fuels for cancer cells. Cancer J 21(2):49–55
Recouvreux MV, Commisso C (2017) Macropinocytosis: A Metabolic adaptation to nutrient stress in cancer. Front Endocrinol (Lausanne) 8:261
Green MR et al (2006) Abraxane, a novel Cremophor-free, albumin-bound particle form of paclitaxel for the treatment of advanced non-small-cell lung cancer. Ann Oncol 17(8):1263–1268
Misale S et al (2019) KRAS G12C NSCLC models are sensitive to direct targeting of KRAS in combination with PI3K inhibition. Clin Cancer Res 25(2):796–807
Jiao J et al (2020) Quicker, deeper and stronger imaging: a review of tumor-targeted, near-infrared fluorescent dyes for fluorescence guided surgery in the preclinical and clinical stages. Eur J Pharm Biopharm 152:123–143
Martelli C et al (2016) Optical imaging probes in oncology. Oncotarget 7(30):48753–48787
Yi X et al (2014) Near-infrared fluorescent probes in cancer imaging and therapy: an emerging field. Int J Nanomedicine 9:1347–1365
Li H et al (2018) Lighting-Up tumor for assisting resection via spraying NIR fluorescent probe of gamma-glutamyltranspeptidas. Front Chem 6:485
Acknowledgements
The authors would like to thank members of the D. Piwnica-Worms laboratory for discussion and suggestions and the Small Animal Imaging Facility of MD Anderson Cancer Center for animal imaging assistance, especially Charles Kingsley and Jorge Delacerda. The Biostatistics Resource, Flow Cytometry Core Facility, and Small Animal Imaging Facility obtain support from NCI Cancer Center Support Grant P30 CA016672.
This work was supported by NIH Grant P50 CA94056 to the MD Anderson Cancer Center–Washington University Inter-Institutional Molecular Imaging Center; the Gerald Dewey Dodd, Jr., Endowed Distinguished Chair of the University of Texas MD Anderson Cancer Center; and a generous gift from the Estate of Barbara Cox Anthony/Koch Foundation. M.N.S. was supported by a Ruth L. Kirchstein Postdoctoral Individual Research Service Award (F32) NCI F32CA250323, the Harold C. and Mary L. Dailey Endowed Fellowship Recognition of Research Excellence (M.D. Anderson Cancer Center), and the Diane Denson Tobola Endowed Fellowship in Ovarian Cancer Research (M.D. Anderson Cancer Center).
Author information
Authors and Affiliations
Contributions
M.N.S. and D.P.W. designed the study. M.N.S., S.T.G., R.M., F.P., B.R., and P.Y. carried out experiments. D.P.W. provided resources and reagents. M.N.S., S.T.G., and D.P.W. analyzed the data, and M.N.S. and D.P.W. wrote the manuscript. All authors edited and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sutton, M.N., Gammon, S.T., Muzzioli, R. et al. RAS-Driven Macropinocytosis of Albumin or Dextran Reveals Mutation-Specific Target Engagement of RAS p.G12C Inhibitor ARS-1620 by NIR-Fluorescence Imaging. Mol Imaging Biol 24, 498–509 (2022). https://doi.org/10.1007/s11307-021-01689-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11307-021-01689-8