Abstract
Introduction
Acute myeloid leukemia (AML) is characterized by a set of malignant proliferations leading to an accumulation of blasts in the bone marrow and blood. The prognosis is pejorative due to the molecular complexity and pathways implicated in leukemogenesis.
Objectives
Our research was focused on comparing the metabolic profiles of leukemic cells in basal culture and deprivation conditions to investigate their behaviors under metabolic stress.
Methods
We performed untargeted metabolomics using 1H HRMAS-NMR. Five human leukemic cell lines—KG1, K562, HEL, HL60 and OCIAML3—were studied in the basal and nutrient deprivation states. A multivariate analysis of the metabolic profile was performed to find over- or under- expressed metabolites in the different cell lines, depending on the experimental conditions.
Results
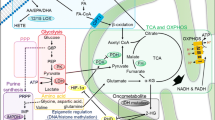
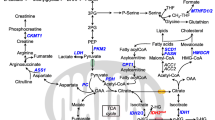
In the basal state, each leukemic cell line exhibited a specific metabolic signature related to the diversity of AML subtypes represented and their phenotypes. When cultured in a serum-free medium, they showed quick metabolic adaptation and continued to proliferate and survive despite the lack of nutrients. Low apoptosis was observed. Increased phosphocholine and glutathione was a common feature of all the observed cell lines, with the maximum increase in these metabolites at 24 h of culture, suggesting the involvement of lipid metabolism and oxidative stress regulators in the survival mechanism developed by the leukemic cells.
Conclusions
Our study provides new insights into the metabolic mechanisms in leukemogenesis and suggests a hierarchy of metabolic pathways activated within leukemic cells, some dependent on their genotypes and others conserved among the subtypes but commonly induced under micro-environmental stress.





Similar content being viewed by others
References
Arber, D. A., et al. (2016). The 2016 revision to the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia. Blood, 127(20), 2391–2405.
Bagnoli, M., et al. (2016). Choline metabolism alteration: A focus on ovarian cancer. Frontiers in Oncology, 6, 153. https://doi.org/10.3389/fonc.2016.00153.
Basquiera, A. L., et al. (2009). Clinical significance of V617F mutation of the JAK2 gene in patients with chronic myeloproliferative disorders. Hematology, 14(6), 323–330. https://doi.org/10.1179/102453309X12473408860226.
Beckonert, O., et al. (2010). High-resolution magic-angle-spinning NMR spectroscopy for metabolic profiling of intact tissues. Nature Protocols, 5(6), 1019–1032. https://doi.org/10.1038/nprot.2010.45.
Bhanot, H., et al. (2015). Pathological glycogenesis through glycogen synthase 1 and suppression of excessive AMP kinase activity in myeloid leukemia cells. Leukemia, 29(7), 1555. https://doi.org/10.1038/leu.2015.46.
Box, J. K., et al. (2016). Nucleophosmin: From structure and function to disease development. BMC Molecular Biology, 17(1), 19. https://doi.org/10.1186/s12867-016-0073-9.
Challen, G. A., et al. (2012). Dnmt3a is essential for hematopoietic stem cell differentiation. Nature Genetics, 44(1), 23–31. https://doi.org/10.1038/ng.1009.
Circu, M. L., & Aw, T. Y. (2008). Glutathione and apoptosis. Free Radical Research, 42(8), 689–706. https://doi.org/10.1080/10715760802317663.
De Braekeleer, E., et al. (2011). ABL1 fusion genes in hematological malignancies: A review. European Journal of Haematology, 86(5), 361–371. https://doi.org/10.1111/j.1600-0609.2011.01586.x.
DeBerardinis, R. J., & Chandel, N. S. (2016). Fundamentals of cancer metabolism. Science. Advances, 2(5), e1600200. https://doi.org/10.1126/sciadv.1600200.
Dewar, B. J., et al. (2010). Metabolic assessment of a novel chronic myelogenous leukemic cell line and an imatinib resistant subline by 1H NMR spectroscopy. Metabolomics, 6(3), 439–450. https://doi.org/10.1007/s11306-010-0204-0.
Döhner, H., et al. (2017). Diagnosis and management of AML in adults: 2017 ELN recommendations from an International Expert Panel. Blood, 129(4), 424–447. https://doi.org/10.1182/blood-2016-08-733196.
Duan-Porter, W. D., et al. (2014). Dynamic conformations of nucleophosmin (NPM1) at a key monomer-monomer interface affect oligomer stability and interactions with granzyme B. PLoS ONE, 9(12), e115062. https://doi.org/10.1371/journal.pone.0115062.
Eskandarpour, M., et al. (2009). Oncogenic NRAS has multiple effects on the malignant phenotype of human melanoma cells cultured in vitro. International Journal of Cancer, 124(1), 16–26. https://doi.org/10.1002/ijc.23876.
Ferret Y et al. (2018) Clinical relevance of IDH1/2 mutant allele burden during follow-up in acute myeloid leukemia. A study by the French ALFA Group. Haematologica, 103(5), 822–829. https://doi.org/10.3324/haematol.2017.183525.
Granata, A., et al. (2013). Choline kinase-alpha by regulating cell aggressiveness and drug sensitivity is a potential druggable target for ovarian cancer. British Journal of Cancer, 110(2), 330. https://doi.org/10.1038/bjc.2013.729.
Han, M., et al. (2019). Epigenetic enzyme mutations: Role in tumorigenesis and molecular inhibitors. Frontiers in Oncology, 9, 194. https://doi.org/10.3389/fonc.2019.00194.
Hernandez-Davies, J. E., et al. (2015). Vemurafenib resistance reprograms melanoma cells towards glutamine dependence. Journal of Translational Medicine, 13(1), 210. https://doi.org/10.1186/s12967-015-0581-2.
Hosseinzadeh, Z., et al. (2012). Down-regulation of the myoinositol transporter SMIT by JAK2. Cellular Physiology and Biochemistry, 30(6), 1473–1480. https://doi.org/10.1159/000343335.
Icard, P., Lincet, H. (2012). A global view of the biochemical pathways involved in the regulation of the metabolism of cancer cells. Biochimica et Biophysica Acta (BBA), 1826(2), 423–433. https://doi.org/10.1016/j.bbcan.2012.07.001.
June, Li, et al. (2006). Nucleophosmin regulates cell cycle progression and stress response in hematopoietic stem/progenitor cells. Journal of Biological Chemistry, 281(24), 16536–16545. https://doi.org/10.1074/jbc.M601386200.
Lash, L., et al. (2015). Glutathione levels and susceptibility to chemically induced injury in two human prostate cancer cell lines. Molecules, 20(6), 10399–10414. https://doi.org/10.3390/molecules200610399.
Markley, J. L., et al. (2017). The future of NMR-based metabolomics. Current Opinion in Biotechnology, 43, 34–40. https://doi.org/10.1016/j.copbio.2016.08.001.
Michalak, K. P., et al. (2015). Key roles of glutamine pathways in reprogramming the cancer metabolism. Oxidative Medicine and Cellular Longevity, 2015, 1–14. https://doi.org/10.1155/2015/964321.
Mondet, J., et al. (2019). Adult patients with de novo acute myeloid leukemia show a functional deregulation of redox balance at diagnosis which is correlated with molecular subtypes and overall survival. Haematologica, 104(9), e393–e397. https://doi.org/10.3324/haematol.2018.206821.
Montalban-Bravo, G., & DiNardo, C. D. (2018). The role of IDH mutations in acute myeloid leukemia. Future Oncology, 14(10), 979–993. https://doi.org/10.2217/fon-2017-0523.
Musharraf, S. G., et al. (2016). Serum metabonomics of acute leukemia using nuclear magnetic resonance spectroscopy. Scientific Reports, 6(1), 30693. https://doi.org/10.1038/srep30693.
Pavlova, N. N., & Thompson, C. B. (2016). The emerging hallmarks of cancer metabolism. Cell Metabolism, 23(1), 27–47. https://doi.org/10.1016/j.cmet.2015.12.006.
Pei, S., et al. (2013). Targeting aberrant glutathione metabolism to eradicate human acute myelogenous leukemia cells. Journal of Biological Chemistry, 288(47), 33542–33558. https://doi.org/10.1074/jbc.M113.511170.
Popovici, C., et al. (1999). The t(6;8)(Q27;P11) translocation in a stem cell myeloproliferative disorder fuses a novel gene, FOP, to fibroblast growth factor receptor. Blood, 93(4), 1381–1389.
Stäubert, C., et al. (2015). Rewired metabolism in drug-resistant leukemia cells: A metabolic switch hallmarked by reduced dependence on exogenous glutamine. Journal of Biological Chemistry, 290(13), 8348–8359. https://doi.org/10.1074/jbc.M114.618769.
Suganuma, K., et al. (2010). Energy metabolism of leukemia cells: Glycolysis versus oxidative phosphorylation. Leukemia and Lymphoma, 51(11), 2112–2119. https://doi.org/10.3109/10428194.2010.512966.
Tiziani, S., et al. (2009). Metabolomic profiling of drug responses in acute myeloid leukaemia cell lines. PLoS ONE, 4(1), e4251. https://doi.org/10.1371/journal.pone.0004251.
Wang, Y., et al. (2013). Rapid diagnosis and prognosis of de novo acute myeloid leukemia by serum metabonomic analysis. Journal of Proteome Research, 12(10), 4393–4401. https://doi.org/10.1021/pr400403p.
Wojtowicz, W., et al. (2018). Serum NMR metabolomics to differentiate haematologic malignancies. Oncotarget, 9(36), 24414. https://doi.org/10.18632/oncotarget.25311.
Ye, M., et al. (2016). FGF21-FGFR1 coordinates phospholipid homeostasis, lipid droplet function, and ER stress in obesity. Endocrinology, 157(12), 4754–4769. https://doi.org/10.1210/en.2016-1710.
Zhang, A., et al. (2013). Cell metabolomics. OMICS: A Journal of Integrative Biology, 17(10), 495–501. https://doi.org/10.1089/omi.2012.0090.
Zhang, X., et al. (2016). Metabolomics profiles delineate uridine deficiency contributes to mitochondria-mediated apoptosis induced by celastrol in human acute promyelocytic leukemia cells. Oncotarget, 7(29), 46557. https://doi.org/10.18632/oncotarget.10286.
Acknowledgements
We thank Michel Bardet, the head of the laboratory of magnetic resonance at CEA Grenoble and Pierre-Alain Bayle, for his invaluable help during the experiments.
Author information
Authors and Affiliations
Contributions
PM conducted the study design; CL pretreated the samples; JM provided help during the stress experiments; FF supervised the metabolomics design of the study; and CL performed the HRMAS-NMR and data analyses. All the authors read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Research involving human and animal participants
This article does not contain any studies with human and/or animal participants.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
11306_2020_1633_MOESM1_ESM.tif
Supplementary file1 Representative metabolites for each cell line: (a) HL60, (b) OCIAML3, (c) HEL, (d) KG1, (e) K562. Statistical analyses (t-tests and ANOVAs) of the NMR peak relative amplitude were performed with GraphPad Prism®. The values are mean ± SE of mean (n=2). (* p ≤ 0.05; ** p ≤ 0.01; *** p ≤ 0.001) (TIF 65973 kb)
11306_2020_1633_MOESM2_ESM.tif
Supplementary file2 Specific regions showing the most representative metabolites for each cell line (e.g. the metabolites with higher levels for each cell line): (a) HEL, (b) KG1, (c) HL60, (d) OCIAML3. For metabolites’ abbreviations, see Table S1 (TIF 55391 kb)
11306_2020_1633_MOESM3_ESM.tif
Supplementary file3 Spectrum section showing the distinction between choline (Cho, 3.20 ppm), phosphocholine (PC, 3.21 ppm) and glycerophosphocholine (GPC, 3.22 ppm) (TIF 67030 kb)
11306_2020_1633_MOESM4_ESM.tif
Supplementary file4 (a) 3D score plot for the HL60 cell line OPLS-DA model (R2Y = 0.94; Q2 = 0.836). (b) 3D score plot for the OCIAML3 cell line OPLS-DA model (R2Y = 0.94; Q2 = 0.818) (TIF 51307 kb)
11306_2020_1633_MOESM5_ESM.docx
Supplementary file5 Metabolites identified with NMR analysis with assigned resonances of detected metabolites in the 1H HRMAS NMR spectra of cells (s singlet, d doublet, dd doublet of doublets, t triplet, q quadruplet, m multiplet). Chemical shifts are relative to the right peak of alanine doublet (1.475 ppm); the peaks used for the univariate statistics are in bold. For asparagine, aspartate, glutamate, taurine and proline, only a part of the bold peak (without any overlapping with either a metabolite or a peak of contaminating compound) was considered. The buckets used in the univariate statistics are given for each metabolite we analyzed (DOCX 14 kb)
Rights and permissions
About this article
Cite this article
Lo Presti, C., Fauvelle, F., Mondet, J. et al. The differential activation of metabolic pathways in leukemic cells depending on their genotype and micro-environmental stress. Metabolomics 16, 13 (2020). https://doi.org/10.1007/s11306-020-1633-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11306-020-1633-z