Abstract
Introduction
Previously constructed Escherichia coli strains that produce 1-propanol use the native threonine pathway, or a heterologous citramalate pathway. However, based on the energy and cofactor requirements of each pathway, a combination of the two pathways produces synergistic effects that increase the theoretical maximum yield with a simultaneous unexplained increase in productivity.
Objective
Identification of key factors that contribute to synergistic effect leading to 1-propanol yield and productivity improvement in E. coli with native threonine pathway and heterologous citramalate pathway.
Method
A combination of snapshot metabolomic profiling and dynamic metabolic turnover analysis were used to identify system-wide perturbations that contribute to the productivity improvement.
Result and Conclusion
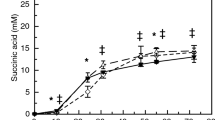
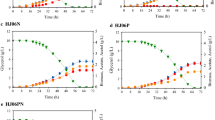
In the presence of both pathways, increased glucose consumption and elevated levels of glycolytic intermediates are attributed to an elevated phosphoenolpyruvate (PEP)/pyruvate ratio that is known to increase the function of the native phosphotransferase. Turnover analysis of nitrogen containing byproducts reveals that ammonia assimilation, required for the threonine pathway, is streamlined when provided with an NAD(P)H surplus in the presence of the citramalate pathway. Our study illustrates the application of metabolomics in identification of factors that alter cellular physiology for improvement of 1-propanol bioproduction.






Similar content being viewed by others
Abbreviations
- GC/MS:
-
Gas chromatography-quadrupole/mass spectrometry
- PCA:
-
Principal component analysis
- LOD:
-
Limit of detection
- LOQ:
-
Limit of quantitation
- PC:
-
Principal component
References
Anesiadis, N., Cluett, W. R., & Mahadevan, R. (2008). Dynamic metabolic engineering for increasing bioprocess productivity. Metabolic Engineering, 10(5), 255–266. https://doi.org/10.1016/j.ymben.2008.06.004.
Atsumi, S., Cann, A. F., Connor, M. R., Shen, C. R., Smith, K. M., Brynildsen, M. P., et al. (2008). Metabolic engineering of Escherichia coli for 1-butanol production. Metabolic Engineering, 10(6), 305–311. https://doi.org/10.1016/j.ymben.2007.08.003.
Atsumi, S., & Liao, J. C. (2008). Directed evolution of Methanococcus jannaschii citramalate synthase for biosynthesis of 1-propanol and 1-butanol by Escherichia coli. Applied and Environmental Microbiology, 74(24), 7802–7808. https://doi.org/10.1128/AEM.02046-08.
Buescher, J. M., Moco, S., Sauer, U., & Zamboni, N. (2010). Ultrahigh performance liquid chromatography—tandem mass spectrometry method for fast and robust quantification of anionic and aromatic metabolites. Analytical Chemistry, 82(11), 4403–4412. https://doi.org/10.1021/ac100101d.
Cann, A. F., & Liao, J. C. (2008). Production of 2-methyl-1-butanol in engineered Escherichia coli. Applied Microbiology and Biotechnology, 81(1), 89–98. https://doi.org/10.1007/s00253-008-1631-y.
Choi, Y. J., Park, J. H., Kim, T. Y., & Lee, S. Y. (2012). Metabolic engineering of Escherichia coli for the production of 1-propanol. Metabolic Engineering, 14(5), 477–486. https://doi.org/10.1016/j.ymben.2012.07.006.
Datsenko, K. A., & Wanner, B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proceedings of the National Academy of Sciences of the United States of America, 97(12), 6640–6645. https://doi.org/10.1073/pnas.120163297.
Fotheringham, I. G., Grinter, N., Pantaleone, D. P., Senkpeil, R. F., & Taylor, P. P. (1999). Engineering of a novel biochemical pathway for the biosynthesis of L-2-aminobutyric acid in Escherichia coli K12. Bioorganic & Medicinal Chemistry, 7(10), 2209–2213. https://doi.org/10.1016/S0968-0896(99)00153-4.
Gold, N., Gowen, C., Lussier, F.-X., Cautha, S., Mahadevan, R., & Martin, V. (2015). Metabolic engineering of a tyrosine-overproducing yeast platform using targeted metabolomics. Microbial Cell Factories, 14(73), 1–16. https://doi.org/10.1186/s12934-015-0252-2.
Hädicke, O., Bettenbrock, K., & Klamt, S. (2015). Enforced ATP futile cycling increases specific productivity and yield of anaerobic lactate production in Escherichia coli. Biotechnology and Bioengineering, 112(10), 2195–2199. https://doi.org/10.1002/bit.25623.
Harada, K., Ohyama, Y., Tabushi, T., Kobayashi, A., & Fukusaki, E. (2008). Quantitative analysis of anionic metabolites for Catharanthus roseus by capillary electrophoresis using sulfonated capillary coupled with electrospray ionization-tandem mass spectrometry. Journal of Bioscience and Bioengineering, 105(3), 249–260. https://doi.org/10.1263/jbb.105.249.
Hasunuma, T., Harada, K., Miyazawa, S. I., Kondo, A., Fukusaki, E., & Miyake, C. (2010). Metabolic turnover analysis by a combination of in vivo 13C-labelling from 13CO2 and metabolic profiling with CE-MS/MS reveals rate-limiting steps of the C3 photosynthetic pathway in Nicotiana tabacum leaves. Journal of Experimental Botany, 61(4), 1041–1051. https://doi.org/10.1093/jxb/erp374.
Hasunuma, T., Sanda, T., Yamada, R., Yoshimura, K., Ishii, J., & Kondo, A. (2011). Metabolic pathway engineering based on metabolomics confers acetic and formic acid tolerance to a recombinant xylose-fermenting strain of Saccharomyces cerevisiae. Microbial Cell Factories, 10(2), 1–13. https://doi.org/10.1186/1475-2859-10-2.
Huffer, S., Roche, C. M., Blanch, H. W., & Clark, D. S. (2012). Escherichia coli for biofuel production: Bridging the gap from promise to practice. Trends in Biotechnology, 30(10), 538–545. https://doi.org/10.1016/j.tibtech.2012.07.002.
Kato, H., Izumi, Y., Hasunuma, T., Matsuda, F., & Kondo, A. (2012). Widely targeted metabolic profiling analysis of yeast central metabolites. Journal of Bioscience and Bioengineering, 113(5), 665–673. https://doi.org/10.1016/j.jbiosc.2011.12.013.
Korneli, C., Bolten, C. J., Godard, T., Franco-Lara, E., & Wittmann, C. (2012). Debottlenecking recombinant protein production in Bacillus megaterium under large-scale conditions—targeted precursor feeding designed from metabolomics. Biotechnology and Bioengineering, 109, 1538–1550. https://doi.org/10.1002/bit.24434.
Liao, J. C., Hou, S. Y., & Chao, Y. P. (1996). Pathway analysis, engineering, and physiological considerations for redirecting central metabolism. Biotechnology and Bioengineering, 52(1), 129–140.
Lommen, A. (2009). MetAlign: Interface-driven, versatile metabolomics tool for hyphenated full-scan mass spectrometry data preprocessing. Analytical Chemistry, 81(8), 3079–3086. https://doi.org/10.1021/ac900036d.
Misko, T. P., Mitchell, W. J., Meadow, N. D., & Roseman, S. (1982). Sugar transport by the bacterial phosphotransferase system. The Journal of Biological Chemistry, 257(23), 14526–14537. https://doi.org/10.1002/pro.5560051020.
Nakayama, Y., Putri, S. P., Bamba, T., & Fukusaki, E. (2014). Metabolic distance estimation based on principle component analysis of metabolic turnover. Journal of Bioscience and Bioengineering, 118(3), 350–355. https://doi.org/10.1016/j.jbiosc.2014.02.014.
Nitta, K., Laviña, W. A., Pontrelli, S., Liao, J. C., Putri, S. P., & Fukusaki, E. (2017). Orthogonal partial least squares/projections to latent structures regression-based metabolomics approach for identification of gene targets for improvement of 1-butanol production in Escherichia coli. Journal of Bioscience and Bioengineering, 124(5), 498–505. https://doi.org/10.1016/j.jbiosc.2017.05.015.
Noguchi, S., Putri, S. P., Lan, E. I., Laviña, W. A., Dempo, Y., Bamba, T., et al. (2016). Quantitative target analysis and kinetic profiling of acyl-CoAs reveal the rate-limiting step in cyanobacterial 1-butanol production. Metabolomics. https://doi.org/10.1007/s11306-015-0940-2.
Ohtake, T., Pontrelli, S., Laviña, W. A., Liao, J. C., Putri, S. P., & Fukusaki, E. (2017). Metabolomics-driven approach to solving a CoA imbalance for improved 1-butanol production in Escherichia coli. Metabolic Engineering, 41(April), 135–143. https://doi.org/10.1016/j.ymben.2017.04.003.
Patnaik, R., Roof, W. D., Young, R. F., & Liao, J. C. (1992). Stimulation of glucose catabolism in Escherichia coli by a potential futile cycle. Journal of Bacteriology, 174(23), 7527–7532. https://doi.org/10.1128/jb.174.23.7527-7532.1992.
Shen, C. R., & Liao, J. C. (2008). Metabolic engineering of Escherichia coli for 1-butanol and 1-propanol production via the keto-acid pathways. Metabolic Engineering, 10(6), 312–320. https://doi.org/10.1016/j.ymben.2008.08.001.
Shen, C. R., & Liao, J. C. (2013). Synergy as design principle for metabolic engineering of 1-propanol production in Escherichia coli. Metabolic Engineering, 17(1), 12–22. https://doi.org/10.1016/j.ymben.2013.01.008.
Solomon, K. V., Moon, T. S., Ma, B., Sanders, T. M., & Prather, K. L. J. (2013). Tuning primary metabolism for heterologous pathway productivity. ACS Synthetic Biology, 2(3), 126–135. https://doi.org/10.1021/sb300055e.
Steffen-Munsberg, F., Vickers, C., Thontowi, A., Schätzle, S., Meinhardt, T., Svedendahl Humble, M., et al. (2013). Revealing the structural basis of promiscuous amine transaminase activity. ChemCatChem, 5(1), 154–157. https://doi.org/10.1002/cctc.201200545.
Teoh, S. T., Putri, S., Mukai, Y., Bamba, T., & Fukusaki, E. (2015). A metabolomics-based strategy for identification of gene targets for phenotype improvement and its application to 1-butanol tolerance in Saccharomyces cerevisiae. Biotechnology for Biofuels. https://doi.org/10.1186/s13068-015-0330-z.
Tsugawa, H., Arita, M., Kanazawa, M., Ogiwara, A., Bamba, T., & Fukusaki, E. (2013). MRMPROBS: A data assessment and metabolite identification tool for large-scale multiple reaction monitoring based widely targeted metabolomics. Analytical Chemistry, 85, 5191–5199. https://doi.org/10.1021/ac400515s.
Tsugawa, H., Tsujimoto, Y., Arita, M., Bamba, T., & Fukusaki, E. (2011). GC/MS based metabolomics: Development of a data mining system for metabolite identification by using soft independent modeling of class analogy (SIMCA). Bioinformatics, 12, 131
Xu, H., Kim, S., Sorek, H., Lee, Y., Jeong, D., Kim, J., et al. (2016). PHO13 deletion-induced transcriptional activation prevents sedoheptulose accumulation during xylose metabolism in engineered Saccharomyces cerevisiae. Metabolic Engineering, 34, 88–96. https://doi.org/10.1016/j.ymben.2015.12.007.
Yoshida, R., Tamura, T., Takaoka, C., Harada, K., Kobayashi, A., Mukai, Y., et al. (2010). Metabolomics-based systematic prediction of yeast lifespan and its application for semi-rational screening of ageing-related mutants. Aging Cell, 9(4), 616–625. https://doi.org/10.1111/j.1474_9726.2010.00590.x.
Acknowledgements
This research was fully supported by Japan Science and Technology (JST), Strategic International Collaborative Research Program, SICORP for JP-US Metabolomics and National Science Foundation (NSF) MCB-1139318.
Author information
Authors and Affiliations
Contributions
SPP designed the study, performed the metabolome analysis and data analysis, and drafted the manuscript. YN performed the turnover analysis and data analysis and helped to draft the manuscript. CS supplied all the strains used in this study, and assisted with the interpretation of metabolome analysis results. SN participated in the metabolome and turnover analysis and data analysis. TB advised the selection of analytical platform, and provided assistance on metabolome analysis and data processing. KN participated in the manuscript preparation, data analysis and data interpretation. SP aided in data interpretation and writing of the manuscript. JL participated in the design of the study, coordination and writing of the manuscript. EF conceived of the study, and participated in its design and coordination. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Research involving human and animal rights
This article does not contain any studies with human participants or animals performed by any of authors.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Putri, S.P., Nakayama, Y., Shen, C. et al. Identifying metabolic elements that contribute to productivity of 1-propanol bioproduction using metabolomic analysis. Metabolomics 14, 96 (2018). https://doi.org/10.1007/s11306-018-1386-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11306-018-1386-0