Abstract
Introduction
Untargeted metabolomics studies for biomarker discovery often have hundreds to thousands of human samples. Data acquisition of large-scale samples has to be divided into several batches and may span from months to as long as several years. The signal drift of metabolites during data acquisition (intra- and inter-batch) is unavoidable and is a major confounding factor for large-scale metabolomics studies.
Objectives
We aim to develop a data normalization method to reduce unwanted variations and integrate multiple batches in large-scale metabolomics studies prior to statistical analyses.
Methods
We developed a machine learning algorithm-based method, support vector regression (SVR), for large-scale metabolomics data normalization and integration. An R package named MetNormalizer was developed and provided for data processing using SVR normalization.
Results
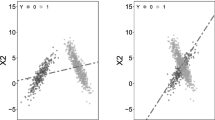
After SVR normalization, the portion of metabolite ion peaks with relative standard deviations (RSDs) less than 30 % increased to more than 90 % of the total peaks, which is much better than other common normalization methods. The reduction of unwanted analytical variations helps to improve the performance of multivariate statistical analyses, both unsupervised and supervised, in terms of classification and prediction accuracy so that subtle metabolic changes in epidemiological studies can be detected.
Conclusion
SVR normalization can effectively remove the unwanted intra- and inter-batch variations, and is much better than other common normalization methods.






Similar content being viewed by others
References
Bijlsma, S., Bobeldijk, L., Verheij, E. R., Ramaker, R., Kochhar, S., Macdonald, I. A., et al. (2006). Large-scale human metabolomics studies: a strategy for data (pre-) processing and validation. Analytical Chemistry, 78(2), 567–574.
Brereton, R. G., & Lloyd, G. R. (2010). Support vector machines for classification and regression. Analyst, 135(2), 230–267.
Burton, L., Ivosev, G., Tate, S., Impey, G., Wingate, J., & Bonner, R. (2008). Instrumental and experimental effects in LC–MS-based metabolomics. Journal of Chromatography B, 871(2), 227–235.
Cairns, D. A., Thompson, D., Perkins, D. N., Stanley, A. J., Selby, P. J., & Banks, R. E. (2008). Proteomic profiling using mass spectrometry—does normalising by total ion current potentially mask some biological differences? Proteomics, 8(1), 21–27.
Cortes, C., & Vapnik, V. (1995). Support-vector networks. Machine Learning, 20, 273–297.
De Livera, A. M., Dias, D. A., De Souza, D., Rupasinghe, T., Pyke, J., Tull, D., et al. (2012). Normalizing and integrating metabolomics data. Analytical Chemistry, 84(24), 10768–10776.
De Livera, A. M., Sysi-Aho, M., Jacob, L., Gagnon-Bartsch, J. A., Castillo, S., Simpson, J. A., et al. (2015). Statistical methods for handling unwanted variation in metabolomics data. Analytical Chemistry, 87(7), 3606–3615.
Dunn, W. B., Broadhurst, D., Begley, P., Zelena, E., Francis-McIntyre, S., Anderson, N., et al. (2011). Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nature Protocols, 6(7), 1060–1083.
Dunn, W. B., Wilson, I. D., Nicholls, A. W., & Broadhurst, D. (2012). The importance of experimental design and QC samples in large-scale and MS-driven untargeted metabolomic studies of humans. Bioanalysis, 4(18), 2249–2264.
Evans, A. M., DeHaven, C. D., Barrett, T., Mitchell, M., & Milgram, E. (2009). Integrated, nontargeted ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small-molecule complement of biological systems. Analytical Chemistry, 81(16), 6656–6667.
FDA. (2013). Guidance for industry, bioanalytical method validation. Food and Drug Administration, Centre for Drug Valuation and Research (CDER).
Fiehn, O. (2002). Metabolomics—the link between genotypes and phenotypes. Plant Molecular Biology, 48(1–2), 155–171.
Fujarewicz, K., Jarzab, M., Eszlinger, M., Krohn, K., Paschke, R., Oczko-Wojciechowska, M., et al. (2007). A multi-gene approach to differentiate papillary thyroid carcinoma from benign lesions: gene selection using support vector machines with bootstrapping. Endocrine-Related Cancer, 14(3), 809–826.
Griffin, J. L., Atherton, H., Shockcor, J., & Atzori, L. (2011). Metabolomics as a tool for cardiac research. Nature Reviews Cardiology, 8(11), 630–643.
Guan, W., Zhou, M., Hampton, C. Y., Benigno, B. B., Walker, L. D., Gray, A., et al. (2009). Ovarian cancer detection from metabolomic liquid chromatography/mass spectrometry data by support vector machines. BMC Bioinformatics, 10, 259.
Huber, W., von Heydebreck, A., Sultmann, H., Poustka, A., & Vingron, M. (2002). Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics, 18(Suppl 1), 96–104.
Kamleh, M. A., Ebbels, T. M. D., Spagou, K., Masson, P., & Want, E. J. (2012). Optimizing the use of quality control samples for signal drift correction in large-scale urine metabolic profiling studies. Analytical Chemistry, 84(6), 2670–2677.
Kuhl, C., Tautenhahn, R., Bottcher, C., Larson, T. R., & Neumann, S. (2012). CAMERA: an integrated strategy for compound spectra extraction and annotation of liquid chromatography/mass spectrometry data sets. Analytical Chemistry, 84(1), 283–289.
Leek, J. T., Scharpf, R. B., Bravo, H. C., Simcha, D., Langmead, B., Johnson, W. E., et al. (2010). Tackling the widespread and critical impact of batch effects in high-throughput data. Nature Reviews Genetics, 11(10), 733–739.
Long, J. Z., Cisar, J. S., Milliken, D., Niessen, S., Wang, C., Trauger, S. A., et al. (2011). Metabolomics annotates ABHD3 as a physiologic regulator of medium-chain phospholipids. Nature Chemical Biology, 7(11), 763–765.
Luan, H. M., Liu, L. F., Meng, N., Tang, Z., Chua, K. K., Chen, L. L., et al. (2015). LC MS-based urinary metabolite signatures in idiopathic Parkinson’s disease. Journal of Proteome Research, 14(1), 467–478.
Lv, H. T., Palacios, G., Hartil, K., & Kurland, I. J. (2011). Advantages of tandem LC–MS for the rapid assessment of tissue-specific metabolic complexity using a pentafluorophenylpropyl stationary phase. Journal of Proteome Research, 10(4), 2104–2112.
Mapstone, M., Cheema, A. K., Fiandaca, M. S., Zhong, X. G., Mhyre, T. R., MacArthur, L. H., et al. (2014). Plasma phospholipids identify antecedent memory impairment in older adults. Nature Medicine, 20(4), 415.
Mayers, J. R., Wu, C., Clish, C. B., Kraft, P., Torrence, M. E., Fiske, B. P., et al. (2014). Elevation of circulating branched-chain amino acids is an early event in human pancreatic adenocarcinoma development. Nature Medicine, 20(10), 1193–1198.
Nicholson, J. K., & Lindon, J. C. (2008). Systems biology—metabonomics. Nature, 455(7216), 1054–1056.
Patti, G. J., Yanes, O., Shriver, L. P., Courade, J. P., Tautenhahn, R., Manchester, M., et al. (2012a). Metabolomics implicates altered sphingolipids in chronic pain of neuropathic origin. Nature Chemical Biology, 8(3), 232–234.
Patti, G. J., Yanes, O., & Siuzdak, G. (2012b). Metabolomics: the apogee of the omics trilogy. Nature Reviews Molecular Cell Biology, 13(4), 263–269.
R Development Core Team. (2015). R: A language and environment for statistical computing. Vienna, Austria. http://www.R-project.org. Accessed 18 June 2015.
Rabinowitz, J. D., & Silhavy, T. J. (2013). Metabolite turns master regulator. Nature, 500(7462), 283–284.
Redestig, H., Fukushima, A., Stenlund, H., Moritz, T., Arita, M., Saito, K., et al. (2009). Compensation for systematic cross-contribution improves normalization of mass spectrometry based metabolomics data. Analytical Chemistry, 81(19), 7974–7980.
Ren, S., Hinzman, A. A., Kang, E. L., Szczesniak, R. D., & Lu, L. J. (2015). Computational and statistical analysis of metabolomics data. Metabolomics, 11(6), 1492–1513.
Rosenberg, L. H., Franzen, B., Auer, G., Lehtio, J., & Forshed, J. (2010). Multivariate meta-analysis of proteomics data from human prostate and colon tumours. BMC Bioinformatics, 11, 468.
Scholz, M., Gatzek, S., Sterling, A., Fiehn, O., & Selbig, J. (2004). Metabolite fingerprinting: detecting biological features by independent component analysis. Bioinformatics, 20(15), 2447–2454.
Smith, C. A., Want, E. J., O’Maille, G., Abagyan, R., & Siuzdak, G. (2006). XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Analytical Chemistry, 78(3), 779–787.
Steinwart, I., & Christmann, A. (2008). Support vector machines. New York: Springer.
Sysi-Aho, M., Katajamaa, M., Yetukuri, L., & Oresic, M. (2007). Normalization method for metabolomics data using optimal selection of multiple internal standards. BMC Bioinformatics, 8, 93.
Tautenhahn, R., Bottcher, C., & Neumann, S. (2008). Highly sensitive feature detection for high resolution LC/MS. BMC Bioinformatics, 9, 504.
van den Berg, R. A., Hoefsloot, H. C., Westerhuis, J. A., Smilde, A. K., & van der Werf, M. J. (2006). Centering, scaling, and transformations: improving the biological information content of metabolomics data. BMC Genomics, 7, 142.
van der Kloet, F. M., Bobeldijk, I., Verheij, E. R., & Jellema, R. H. (2009). Analytical error reduction using single point calibration for accurate and precise metabolomic phenotyping. Journal of Proteome Research, 8(11), 5132–5141.
Veselkov, K. A., Vingara, L. K., Masson, P., Robinette, S. L., Want, E., Li, J. V., et al. (2011). Optimized preprocessing of ultra-performance liquid chromatography/mass spectrometry urinary metabolic profiles for improved information recovery. Analytical Chemistry, 83(15), 5864–5872.
Wang, S. Y., Kuo, C. H., & Tseng, Y. F. J. (2013). Batch normalizer: a fast total abundance regression calibration method to simultaneously adjust batch and injection order effects in liquid chromatography/time-of-flight mass spectrometry-based metabolomics data and comparison with current calibration methods. Analytical Chemistry, 85(2), 1037–1046.
Wang, T. J., Larson, M. G., Vasan, R. S., Cheng, S., Rhee, E. P., McCabe, E., et al. (2011). Metabolite profiles and the risk of developing diabetes. Nature Medicine, 17(4), 448–453.
Wang, W. X., Zhou, H. H., Lin, H., Roy, S., Shaler, T. A., Hill, L. R., et al. (2003). Quantification of proteins and metabolites by mass spectrometry without isotopic labeling or spiked standards. Analytical Chemistry, 75(18), 4818–4826.
Weiss, R. H., & Kim, K. M. (2012). Metabolomics in the study of kidney diseases. Nature Reviews Nephrology, 8(1), 22–33.
Acknowledgments
The work is financially supported by the funding from Interdisciplinary Research Center on Biology and Chemistry (IRCBC), Chinese Academy of Sciences (CAS), and the National Natural Science Foundation of China (Grants 21575151 and 81573246). Z.-J. Z. is supported by Thousand Youth Talents Program (The Recruitment Program of Global Youth Experts from Chinese government). This work is also partially supported by Agilent Technologies Thought Leader Award.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing financial interest.
Ethical approval
All institutional and national guidelines for the care and use of biological samples were followed. The data acquired were in accordance with appropriate ethical requirements.
Research involving human participants
The human study was approved by the ethics committee of Shandong Cancer Hospital affiliated to Shandong University, Shandong Province, China.
Informed consent
All written informed consents were obtained from all participants involved in this study.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Shen, X., Gong, X., Cai, Y. et al. Normalization and integration of large-scale metabolomics data using support vector regression. Metabolomics 12, 89 (2016). https://doi.org/10.1007/s11306-016-1026-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11306-016-1026-5