Abstract
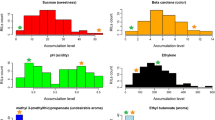
Elucidating the determinants of tomato nutritional value and fruit quality to introduce improved varieties on the international market represents a major challenge for crop biotechnology. Different strategies can be undertaken to exploit the natural variability of Solanum to re-incorporate lost allelic diversity into commercial varieties. One of them is the characterization of selected germplasm for breeding programs. To achieve this goal, 18 RILs (S. lycopersicum × S. pimpinellifolium) were comprehensively phenotyped for fruit polar metabolites and quality associated traits. Metabolites were quantified by GC–MS and 1H NMR. Integrative analyses by neuronal clustering and network construction revealed that fruit properties are strongly associated with the metabolites aspartate, serine, glutamate and 2-oxoglutarate. Shelf life and firmness appeared to be linked to malate content. By a comparative analysis of the whole data set, ten RILs presented higher number of traits with positive effect than the S. lycopersicum × S. pimpinellifolium hybrid. Thus, these lines can be proposed as promising candidates for breeding programs aimed to improve fruit quality.





Similar content being viewed by others
References
Abriata, L. A. (2012). Utilization of NMR spectroscopy to study biological fluids and metabolic processes: two introductory activities. Concepts in Magnetic Resonance Part A, 40A(4), 171–178. doi:10.1002/cmr.a.21235.
Alba, R., Payton, P., Fei, Z., McQuinn, R., Debbie, P., Martin, G. B., et al. (2005). Transcriptome and selected metabolite analyses reveal multiple points of ethylene control during tomato fruit development. Plant Cell, 17(11), 2954–2965. doi:10.1105/tpc.105.036053.
Araújo, W. L., Tohge, T., Osorio, S., Lohse, M., Balbo, I., Krahnert, I., et al. (2012). Antisense inhibition of the 2-oxoglutarate dehydrogenase complex in tomato demonstrates its importance for plant respiration and during leaf senescence and fruit maturation. Plant Cell, 24(6), 2328–2351. doi:10.1105/tpc.112.099002.
Asano, N., Kato, A., Matsui, K., Watson, A. A., Nash, R. J., Molyneux, R. J., et al. (1997). The effects of calystegines isolated from edible fruits and vegetables on mammalian liver glycosidases. Glycobiology, 7(8), 1085–8. http://www.ncbi.nlm.nih.gov/pubmed/9455909.
Bai, Y., & Lindhout, P. (2007). Domestication and breeding of tomatoes: what have we gained and what can we gain in the future? Annals of Botany, 100(5), 1085–1094. doi:10.1093/aob/mcm150.
Bekkouche, K., Daali, Y., Cherkaoui, S., Veuthey, J. L., & Christen, P. (2001). Calystegine distribution in some Solanaceous species. Phytochemistry, 58(3), 455–62. http://www.ncbi.nlm.nih.gov/pubmed/11557078.
Bermúdez, L., de Godoy, F., Baldet, P., Demarco, D., Osorio, S., Quadrana, L., et al. (2014). Silencing of the tomato sugar partitioning affecting protein (SPA) modifies sink strength through a shift in leaf sugar metabolism. The Plant Journal, 77(5), 676–687. doi:10.1111/tpj.12418.
Bermúdez, L., Urias, U., Milstein, D., Kamenetzky, L., Asis, R., Fernie, A. R., et al. (2008). A candidate gene survey of quantitative trait loci affecting chemical composition in tomato fruit. Journal of Experimental Botany, 59(10), 2875–2890. doi:10.1093/jxb/ern146.
Bretó, M. P., Asins, M. J., & Carbonell, E. A. (1993). Genetic variability in Lycopersicon species and their genetic relationships. Theoretical and Applied Genetics,. doi:10.1007/BF00223815.
Bucheli, P., Voirol, E., de la Torre, R., López, J., Rytz, a, Tanksley, S. D., & Pétiard, V. (1999). Definition of nonvolatile markers for flavor of tomato (Lycopersicon esculentum Mill.) as tools in selection and breeding. Journal of Agricultural and Food Chemistry, 47(2), 659–64. http://www.ncbi.nlm.nih.gov/pubmed/10563949.
Carnevillier, V., Schlich, P., Guerreau, J., Charpentier, C., & Feuillat, M. (1999). Characterization of the production regions of Chardonnay wines by analysis of free amino acids. Vitis, 38(1), 37–42.
Carreno-Quintero, N., Bouwmeester, H. J., & Keurentjes, J. J. B. (2013). Genetic analysis of metabolome-phenotype interactions: from model to crop species. Trends in Genetics, 29(1), 41–50. doi:10.1016/j.tig.2012.09.006.
Causse, M., Duffe, P., Gomez, M. C., Buret, M., Damidaux, R., Zamir, D., et al. (2004). A genetic map of candidate genes and QTLs involved in tomato fruit size and composition. Journal of Experimental Botany, 55(403), 1671–1685. doi:10.1093/jxb/erh207.
Causse, M., Saliba-Colombani, V., Lecomte, L., Duffé, P., Rousselle, P., & Buret, M. (2002). QTL analysis of fruit quality in fresh market tomato: a few chromosome regions control the variation of sensory and instrumental traits. Journal of Experimental Botany, 53(377), 2089–2098. doi:10.1093/jxb/erf058.
Centeno, D. C., Osorio, S., Nunes-Nesi, A., Bertolo, A. L. F., Carneiro, R. T., Araújo, W. L., et al. (2011). Malate plays a crucial role in starch metabolism, ripening, and soluble solid content of tomato fruit and affects postharvest softening. Plant Cell, 23(1), 162–184. doi:10.1105/tpc.109.072231.
Di Rienzo, J. A., Casanoves, F., Balzarini, M. G., Gonzalez, L., Tablada, M., & Robledo, C. W. (2011). InfoStat version 2011. http://www.infostat.com.ar.
Fernandez-Ruiz, V., Sanchez-Mata, M. C., Camara, M., Torija, M. E., Chaya, C., Galiana-Balaguer, L., et al. (2004). Internal quality characterization of fresh tomato fruits. HortScience, 39(2), 339–345. Retrieved July 1, 2013 from http://hortsci.ashspublications.org/content/39/2/339.abstract.
Fernie, A. R., & Schauer, N. (2009). Metabolomics-assisted breeding: a viable option for crop improvement? Trends in Genetics, 25(1), 39–48. doi:10.1016/j.tig.2008.10.010.
Fitzpatrick, T. B., Basset, G. J. C., Borel, P., Carrari, F., DellaPenna, D., Fraser, P. D., et al. (2012). Vitamin deficiencies in humans: can plant science help? Plant Cell, 24(2), 395–414. doi:10.1105/tpc.111.093120.
Foolad, M. R. (2007). Genome mapping and molecular breeding of tomato. International Journal of Plant Genomics, 2007, 64358. doi:10.1155/2007/64358.
Foolad, M. R., Chen, F. Q., & Lin, G. Y. (1998). RFLP mapping of QTLs conferring salt tolerance during germination in an interspecific cross of tomato. Theoretical and Applied Genetics, 97(7), 1133–1144. doi:10.1007/s001220051002.
Fulton, T. M., Beck-Bunn, T., Emmatty, D., Eshed, Y., Lopez, J., Petiard, V., et al. (1997). QTL analysis of an advanced backcross of Lycopersicon peruvianum to the cultivated tomato and comparisons with QTLs found in other wild species. Theoretical and Applied Genetics, 95(5–6), 881–894. doi:10.1007/s001220050639.
Fulton, T. M., Grandillo, S., Beck-Bunn, T., Fridman, E., Frampton, A., Lopez, J., et al. (2000). Advanced backcross QTL analysis of a Lycopersicon esculentum × Lycopersicon parviflorum cross. Theoretical and Applied Genetics, 100(7), 1025–1042. doi:10.1007/s001220051384.
Gallo, M., Rodríguez, G. R., Zorzoli, R., & Pratta, G. R. (2011). Ligamiento entre caracteres cuantitativos de calidad de fruto y perfiles polipeptídicos del pericarpio en dos estados de madurez en tomate. Revista de la Facultad de Ciencias Agrarias Universidad Nacional de Cuyo, 43(2), 145–156.
Gärtner, T., Steinfath, M., Andorf, S., Lisec, J., Meyer, R. C., Altmann, T., et al. (2009). Improved heterosis prediction by combining information on DNA- and metabolic markers. PLoS ONE, 4(4), e5220. doi:10.1371/journal.pone.0005220.
Gechev, T. S., Hille, J., Woerdenbag, H. J., Benina, M., Mehterov, N., Toneva, V., et al. (2014). Natural products from resurrection plants: potential for medical applications. Biotechnology Advances, 32(6), 1091–1101. doi:10.1016/j.biotechadv.2014.03.005.
Giovannoni, J. (2001). Molecular biology of fruit maturation and ripening. Annual Review of Plant Physiology and Plant Molecular Biology, 52, 725–749. doi:10.1146/annurev.arplant.52.1.725.
Goff, S. A., & Klee, H. J. (2006). Plant volatile compounds: sensory cues for health and nutritional value? Science, 311(5762), 815–819. doi:10.1126/science.1112614.
Goulet, C., Mageroy, M. H., Lam, N. B., Floystad, A., Tieman, D. M., & Klee, H. J. (2012). Role of an esterase in flavor volatile variation within the tomato clade. Proceedings of the National Academy of Sciences of the United States of America, 109(46), 19009–19014. doi:10.1073/pnas.1216515109.
Guimerà, R., & Nunes Amaral, L. A. (2005). Functional cartography of complex metabolic networks. Nature, 433(7028), 895–900. doi:10.1038/nature03288.
Hall, R. D., Brouwer, I. D., & Fitzgerald, M. A. (2008). Plant metabolomics and its potential application for human nutrition. Physiologia Plantarum, 132(2), 162–175. doi:10.1111/j.1399-3054.2007.00989.x.
Hermann, A., & Schauer, N. (2013). Metabolomics-assisted plant breeding. In Wolfram Weckwerth & Guenter Kahl (Eds.), Handbook of Plant Metabolomics (pp. 247–254). John: Wiley & Sons.
Hu, C., Shi, J., Quan, S., Cui, B., Kleessen, S., Nikoloski, Z., et al. (2014). Metabolic variation between japonica and indica rice cultivars as revealed by non-targeted metabolomics. Scientific Reports, 4, 5067. doi:10.1038/srep05067.
Jocković, N., Fischer, W., Brandsch, M., Brandt, W., & Dräger, B. (2013). Inhibition of human intestinal α-glucosidases by calystegines. Journal of Agricultural and Food Chemistry, 61(23), 5550–5557. doi:10.1021/jf4010737.
Kamenetzky, L., Asís, R., Bassi, S., de Godoy, F., Bermúdez, L., Fernie, A. R., et al. (2010). Genomic analysis of wild tomato introgressions determining metabolism- and yield-associated traits. Plant Physiology, 152(4), 1772–1786. doi:10.1104/pp.109.150532.
Kind, T., Wohlgemuth, G., Lee, D. Y., Lu, Y., Palazoglu, M., Shahbaz, S., et al. (2009). FiehnLib—mass spectral and retention index libraries for metabolomics based on quadrupole and time-of-flight gas chromatography/mass spectrometry. Analytical Chemistry, 81(24), 10038–10048.
Kisaka, H., Kida, T., & Miwa, T. (2006). Antisense suppression of glutamate decarboxylase in tomato (Lycopersicon esculentum L.) results in accumulation of glutamate in transgenic tomato fruits. Plant Biotechnology, 274, 267–274.
Klee, H. J. (2013). Purple tomatoes: longer lasting, less disease, and better for you. Current Biology, 23(12), R520–R521. doi:10.1016/j.cub.2013.05.010.
Kopka, J., Schauer, N., Krueger, S., Birkemeyer, C., Usadel, B., Bergmuller, E., et al. (2005). GMD@CSB.DB: the Golm Metabolome Database. Bioinformatics, 21(8), 1635–1638.
Kvasnicka, F., Jockovic, N., Dräger, B., Sevcík, R., Cepl, J., & Voldrich, M. (2008). Electrophoretic determination of calystegines A3 and B2 in potato. Journal of Chromatography A, 1181(1–2), 137–144. doi:10.1016/j.chroma.2007.12.037.
Lee, J. M., Joung, J.-G., McQuinn, R., Chung, M.-Y., Fei, Z., Tieman, D., et al. (2012). Combined transcriptome, genetic diversity and metabolite profiling in tomato fruit reveals that the ethylene response factor SlERF6 plays an important role in ripening and carotenoid accumulation. The Plant Journal, 70(2), 191–204. doi:10.1111/j.1365-313X.2011.04863.x.
Liberatti, D., Rodriguez, G., Zorzoli, R., & Pratta, G. R. (2013). Tomato second cycle hybrids differ from parents at three levels of genetic variation. International Journal of Plant Breeding, 7(1), 1–7. http://www.globalsciencebooks.info/JournalsSup/images/Sample/IJPB_7(1)1-6o.pdf.
Lin, T., Zhu, G., Zhang, J., Xu, X., Yu, Q., Zheng, Z., et al. (2014). Genomic analyses provide insights into the history of tomato breeding. Nature Genetics, 46(11), 1220–1226. doi:10.1038/ng.3117.
Lisec, J., Römisch-Margl, L., Nikoloski, Z., Piepho, H.-P., Giavalisco, P., Selbig, J., et al. (2011). Corn hybrids display lower metabolite variability and complex metabolite inheritance patterns. The Plant Journal, 68(2), 326–336. doi:10.1111/j.1365-313X.2011.04689.x.
Lisec, J., Schauer, N., Kopka, J., Willmitzer, L., & Fernie, A. R. (2006). Gas chromatography mass spectrometry-based metabolite profiling in plants. Nature Protocols, 1(1), 387–396. doi:10.1038/nprot.2006.59.
Lisec, J., Steinfath, M., Meyer, R. C., Selbig, J., Melchinger, A. E., Willmitzer, L., & Altmann, T. (2009). Identification of heterotic metabolite QTL in Arabidopsis thaliana RIL and IL populations. The Plant Journal, 59(5), 777–788. doi:10.1111/j.1365-313X.2009.03910.x.
Luedemann, A., Strassburg, K., Erban, A., & Kopka, J. (2008). TagFinder for the quantitative analysis of gas chromatography–mass spectrometry (GC-MS)-based metabolite profiling experiments. Bioinformatics, 24(5), 732–737. doi:10.1093/bioinformatics/btn023.
Mattoo, A. K., Sobolev, A. P., Neelam, A., Goyal, R. K., Handa, A. K., & Segre, A. L. (2006). Nuclear magnetic resonance spectroscopy-based metabolite profiling of transgenic tomato fruit engineered to accumulate spermidine and spermine reveals enhanced anabolic and nitrogen-carbon interactions. Plant Physiology, 142(4), 1759–1770. doi:10.1104/pp.106.084400.
Miller, J. C., & Tanksley, S. D. (1990). RFLP analysis of phylogenetic relationships and genetic variation in the genus Lycopersicon. Theoretical and Applied Genetics,. doi:10.1007/BF00226743.
Milone, D., Stegmayer, G., Gerard, M., Kamenetzky, L., López, M., & Carrari, F. (2010). Métodos de agrupamiento no supervisado para la integración de datos genómicos y metabólicos de múltiples líneas de introgresión. Inteligencia Artificial, 13(44), 56–66. doi:10.4114/ia.v13i44.1046.
Moose, S. P., & Mumm, R. H. (2008). Molecular plant breeding as the foundation for 21st century crop improvement. Plant Physiology, 147(3), 969–977. doi:10.1104/pp.108.118232.
Mulas, G., Galaffu, M. G., Pretti, L., Nieddu, G., Mercenaro, L., Tonelli, R., & Anedda, R. (2011). NMR analysis of seven selections of vermentino grape berry: metabolites composition and development. Journal of Agricultural and Food Chemistry, 59(3), 793–802. doi:10.1021/jf103285f.
Niggeweg, R., Michael, A. J., & Martin, C. (2004). Engineering plants with increased levels of the antioxidant chlorogenic acid. Nature Biotechnology, 22(6), 746–754. doi:10.1038/nbt966.
Pascual, L., Desplat, N., Huang, B. E., Desgroux, A., Bruguier, L., Bouchet, J.-P., et al. (2014). Potential of a tomato MAGIC population to decipher the genetic control of quantitative traits and detect causal variants in the resequencing era. Plant Biotechnology Journal,. doi:10.1111/pbi.12282.
Paterson, A. H., Lander, E. S., Hewitt, J. D., Peterson, S., Lincoln, S. E., & Tanksley, S. D. (1988). Resolution of quantitative traits into Mendelian factors by using a complete linkage map of restriction fragment length polymorphisms. Nature, 335(6192), 721–726. doi:10.1038/335721a0.
Pratta, G. R., Rodríguez, G. R., Zorzoli, R., Picardi, L. A., & Valle, E. M. (2011a). Biodiversity in a tomato germplasm for free amino acid and pigment content of ripening fruits. American Journal of Plant Sciences, 02(02), 255–261. doi:10.4236/ajps.2011.22027.
Pratta, G. R., Rodriguez, G. R., Zorzoli, R., Valle, E. M., & Picardi, L. A. (2011b). Phenotypic and molecular characterization of selected tomato recombinant inbred lines derived from the cross Solanum lycopersicum × S. pimpinellifolium. Journal of Genetics, 90(2), 229–37. http://www.ncbi.nlm.nih.gov/pubmed/21869471.
Price, A. H. (2006). Believe it or not, QTLs are accurate! Trends in Plant Science, 11(5), 213–216. doi:10.1016/j.tplants.2006.03.006.
Rambla, J. L., Tikunov, Y. M., Monforte, A. J., Bovy, A. G., & Granell, A. (2014). The expanded tomato fruit volatile landscape. Journal of Experimental Botany, 65(16), 4613–4623. doi:10.1093/jxb/eru128.
Rao, J., Cheng, F., Hu, C., Quan, S., & Lin, H. (2014). Metabolic map of mature maize kernels. Metabolomics,. doi:10.1007/s11306-014-0624-3.
Riedelsheimer, C., Czedik-Eysenberg, A., Grieder, C., Lisec, J., Technow, F., Sulpice, R., et al. (2012). Genomic and metabolic prediction of complex heterotic traits in hybrid maize. Nature Genetics, 44(2), 217–220. doi:10.1038/ng.1033.
Rodríguez, G. R., Pratta, G. R., Zorzoli, R., Picardi, L. A., & Divergent-antagonistic, L. A. (2006). Recombinant lines obtained from an interspecific cross between Lycopersicon species selected by fruit weight and fruit shelf life. Journal of the American Society for Horticultural Science, 131(5), 651–656.
Roessner, U., Luedemann, A., Brust, D., Fiehn, O., Linke, T., Willmitzer, L., & Fernie, A. (2001). Metabolic profiling allows comprehensive phenotyping of genetically or environmentally modified plant systems. Plant Cell, 13(1), 11–29.
Roessner-Tunali, U., Lytovchenko, A., Carrari, F., Bruedigam, C., Granot, D., & Fernie, A. R. (2003). Metabolic profiling of transgenic tomato plants overexpressing hexokinase reveals that the influence of hexose phosphorylation diminishes during fruit development. Plant Physiology, 133(2), 84–99. doi:10.1104/pp.103.023572.84.
Saeed, A. I., Bhagabati, N. K., Braisted, J. C., Liang, W., Sharov, V., Howe, E. A., et al. (2006). TM4 microarray software suite. Methods in Enzymology, 411, 134–93. doi:10.1016/S0076-6879(06)11009-5
Saito, K., & Matsuda, F. (2010). Metabolomics for functional genomics, systems biology, and biotechnology. Annual Review of Plant Biology, 61, 463–489. doi:10.1146/annurev.arplant.043008.092035.
Sato, S., Tabata, S., Hirakawa, H., Asamizu, E., Shirasawa, K., Isobe, S., et al. (2012). The tomato genome sequence provides insights into fleshy fruit evolution. Nature, 485(7400), 635–641. doi:10.1038/nature11119.
Sauvage, C., Segura, V., Bauchet, G., Stevens, R., Do, P. T., Nikoloski, Z., et al. (2014). Genome-Wide association in tomato reveals 44 candidate loci for fruit metabolic traits. Plant Physiology, 165(3), 1120–1132. doi:10.1104/pp.114.241521.
Schauer, N., Semel, Y., Balbo, I., Steinfath, M., Repsilber, D., Selbig, J., et al. (2008). Mode of inheritance of primary metabolic traits in tomato. Plant Cell, 20(3), 509–523. doi:10.1105/tpc.107.056523.
Schauer, N., Zamir, D., & Fernie, A. R. (2005). Metabolic profiling of leaves and fruit of wild species tomato: a survey of the Solanum lycopersicum complex. Journal of Experimental Botany, 56(410), 297–307. doi:10.1093/jxb/eri057.
Semel, Y., Nissenbaum, J., Menda, N., Zinder, M., Krieger, U., Issman, N., et al. (2006). Overdominant quantitative trait loci for yield and fitness in tomato. Proceedings of the National Academy of Sciences of the United States of America, 103(35), 12981–12986. doi:10.1073/pnas.0604635103.
Sorrequieta, A., Abriata, L., Boggio, S., & Valle, E. (2013). Off-the-vine ripening of tomato fruit causes alteration in the primary metabolite composition. Metabolites, 3(4), 967–978. doi:10.3390/metabo3040967.
Stegmayer, G., Milone, D., Kamenetzky, L., Lopez, M., & Carrari, F. (2009). Neural network model for integration and visualization of introgressed genome and metabolite data. International Joint Conference on Neural Networks, 2009, 2983–2989. doi:10.1109/IJCNN.2009.5179039.
Swamy, B. P. M., & Sarla, N. (2008). Yield-enhancing quantitative trait loci (QTLs) from wild species. Biotechnology Advances, 26(1), 106–120. doi:10.1016/j.biotechadv.2007.09.005.
Tieman, D., Bliss, P., McIntyre, L. M., Blandon-Ubeda, A., Bies, D., Odabasi, A. Z., et al. (2012). The chemical interactions underlying tomato flavor preferences. Current Biology, 22(11), 1035–1039. doi:10.1016/j.cub.2012.04.016.
Tigchelaar, E. C. (1986). Breeding vegetable crops (M. J. Basset, Ed.) (pp. 135–170). Westport, CT: AVI Publishing Company.
Wahyuni, Y., Ballester, A.-R., Tikunov, Y., Vos, R. C. H., Pelgrom, K. T. B., Maharijaya, A., et al. (2012). Metabolomics and molecular marker analysis to explore pepper (Capsicum sp.) biodiversity. Metabolomics, 9(1), 130–144. doi:10.1007/s11306-012-0432-6.
Warren, G. F. (1998). Spectacular increases in crop yields in the United States in the twentieth century. Weed Technology, 12(4), 752–760.
Willcox, J. K., Catignani, G. L., & Lazarus, S. (2003). Tomatoes and cardiovascular health. Critical Reviews in Food Science and Nutrition, 43(1), 1–18. doi:10.1080/10408690390826437.
Yang, W., Bai, X., Kabelka, E., Eaton, C., Kamoun, S., Knaap, E. Van, & Van Der Francis, E. (2004). Discovery of single nucleotide polymorphisms in Lycopersicon esculentum by computer aided analysis of expressed sequence tags. Molecular Breeding, 14(1), 21–34.
Zorzoli, R., Pratta, G. R., & Picardi, L. A. (2000). Variabilidad genética para la vida postcosecha y el peso de los frutos en tomate para familias F3 de un híbrido interespecífico. Pesquisa Agropecuária Brasileira, 35(12), 2423–2427. doi:10.1590/S0100-204X2000001200013.
Acknowledgments
M.G. López was recipient of a fellowship of Consejo Nacional de Investigaciones Científicas y Técnicas (Argentina). This work was partially supported with grants from Instituto Nacional de Tecnología Agropecuaria, Consejo Nacional de Investigaciones Científicas y Técnicas, Agencia Nacional de Promoción Científica y Tecnológica (Argentina) and from the Max Planck Society (Germany).
Disclosures
This work was carried out in compliance with current laws governing genetic experimentation in Argentina. The authors declared no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
11306_2015_798_MOESM1_ESM.xls
Supplementary Tables List of properties for metabolite (60) determination (Table S1). Relative values of mature fruit metabolic contents (60) and agronomic (14) trait from all the material analyzed are provided (Table S2). Details of the components of each neuron in the *omeSOM 9x9 map are also presented (Table S3) (XLS 294 kb)
11306_2015_798_MOESM2_ESM.tif
Supplementary Fig. S1 Representative 1H-NMR spectrum of tomato pericarp extract in buffered D2O. Known compounds are annotated according to Table S1. The 1H chemical shifts used for identification/quantification of the nineteen metabolites were determined at pH 7.4 and expressed as relative values to that of TSP at 0 ppm. Ala: alanine (doublet at 1.45 ppm), Asn: asparagine (multiplet at 2.82), Asp: aspartate (doublet of doublet ar 2.76), Citrate (doublet of doublets at 2.51), Ethanol (triplet at 1.15), Frc: fructose (multiplet at 4.08), GABA (multiplet at 1.84), Glc: glucose (doublet at 4.62), Glu: glutamate (multiplet at 2.05), Gln: glutamine (multiplet at 2.44), Ile: Isoleucine (doublet at 0.98), Malate (doublet of doublets at 4.27), Methanol (singlet at 3.31), Phe:phenylalanine (multiplet at 7.38), Pyr: Pyruvate (singlet at 2.35), Suc: sucrose (doublet at 5.38), Thr: Threonine (doublet at 1.3), Trp: Tryptophan (doublet ar 7.51), Val: valine (doublet at 1.01) (TIFF 40048 kb)
11306_2015_798_MOESM3_ESM.tif
Supplementary Fig. S2 Values of relative distance between the same metabolite (16) measured by GC-MS and 1H-NMR across different map sizes evaluated are plotted (TIFF 1132 kb)
11306_2015_798_MOESM4_ESM.tif
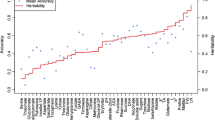
Supplementary Fig. S3 Different mode of inheritance of metabolic (60) and agronomic (14) traits. Traits (metabolic –blue bars- and agronomic –red bars-) were classified according their mode of inheritance following the analysis proposed by Lisec et al. (2011) (see Material and Method section) (TIFF 2033 kb)
Rights and permissions
About this article
Cite this article
López, M.G., Zanor, M.I., Pratta, G.R. et al. Metabolic analyses of interspecific tomato recombinant inbred lines for fruit quality improvement. Metabolomics 11, 1416–1431 (2015). https://doi.org/10.1007/s11306-015-0798-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11306-015-0798-3