Abstract
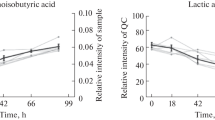
Metabolomics is a growing research field where new protocols are rapidly developed and new applications discovered. Common applications include biomarker discovery and elucidation of drug metabolism. However, the development of such protocols rarely includes a systematic optimization followed by validation with real samples. Here a GC/MS-based protocol using methoximation followed by silylation with N-tert-butyldimethylsilyl-N-methyltrifluoroacetamide (MTBSTFA) for analysis of blood plasma metabolites is thoroughly developed and optimized from derivatization to detection with statistical design of experiments (DOE). Validation was performed with blood plasma samples and proved the methodology to be efficient, rapid and reliable with a total of 51 analyses performed in 24 h, with linear responses, low detection limits and good precision. The obtained chromatograms were much cleaner, due to the absence of glucose overloading, and the data was found to drift less with MTBSTFA derivatisation than with MTBSTFA derivatisation.






Similar content being viewed by others
References
Araujo, P. (2009). Key aspects of analytical method validation and linearity evaluation. Journal of Chromatography B, 877, 2224–2234.
Araujo, P. W., & Brereton, R. G. (1996a). Experimental design I. Screening. Trends in Analytical Chemistry, 15, 26–31.
Araujo, P. W., & Brereton, R. G. (1996b). Experimental design II. Optimization. Trends in Analytical Chemistry, 15, 63–70.
Araujo, P. W., & Brereton, R. G. (1996c). Experimental design III. Quantification. Trends in Analytical Chemistry, 15, 156–163.
Asres, D. D., & Perreault, H. (1997). Monosaccharide permethylation products for gas chromatography - mass spectrometry: How reaction conditions can influence isomeric ratios. Canadian Journal of Chemistry, 75, 1385–1392.
Begley, P., Francis-McIntyre, S., Dunn, W. B., et al. (2009). Development and performance of a gas chromatography-time-of-flight mass spectrometry analysis for large-scale nontargeted metabolomic studies of human serum. Analytical Chemistry, 81, 7038–7046.
Birkemeyer, C., Kolasa, A., & Kopka, J. (2003). Comprehensive chemical derivatization for gas chromatography-mass spectrometry-based multi-targeted profiling of the major phytohormones. Journal of Chromatography A, 993, 89–102.
Büscher, J. M., Czernik, D., Ewald, J. C., Sauer, U., & Zamboni, N. (2009). Cross-platform comparison of methods for quantitative metabolomics of primary metabolism. Analytical Chemistry, 81, 2135–2143.
Chorell, E., Moritz, T., Branth, S., Antti, H., & Svensson, M. B. (2009). Predictive metabolomics evaluation of nutrition-modulated metabolic stress responses in human blood serum during the early recovery phase of strenuous physical exercise. Journal of Proteome Research, 8, 2966–2977.
Cozzolino, D., Flood, L., Bellon, J., Gishen, M., & Lopes, M. D. B. (2006). Combining near infrared spectroscopy and multivariate analysis as a tool to differentiate different strains of Saccharomyces cerevisiae: A metabolomic study. Yeast, 23, 1089–1096.
Danielsson, A. P. H., Moritz, T., Mulder, H., & Spégel, P. (2010). Development and optimization of a metabolomic method for analysis of adherent cell cultures. Analytical Biochemistry, 404, 30–39.
Ewald, J. C., Heux, S. p., & Zamboni, N. (2009). High-throughput quantitative metabolomics: Workflow for cultivation, quenching, and analysis of yeast in a multiwell format. Analytical Chemistry, 81, 3623–3629.
Fiehn, O. (2002). Metabolomics—the link between genotypes and phenotypes. Plant Molecular Biology, 48, 155–171.
Fiehn, O. (2008). Extending the breadth of metabolite profiling by gas chromatography coupled to mass spectrometry. Trends in Analytical Chemistry, 27, 261–269.
Fiehn, O., Kopka, J., Trethewey, R. N., & Willmitzer, L. (2000). Identification of uncommon plant metabolites based on calculation of elemental compositions using gas chromatography and quadrupole mass spectrometry. Analytical Chemistry, 72, 3573–3580.
Gullberg, J., Jonsson, P., Nordström, A., Sjöström, M., & Moritz, T. (2004). Design of experiments: An efficient strategy to identify factors influencing extraction and derivatization of Arabidopsis thaliana samples in metabolomic studies with gas chromatography/mass spectrometry. Analytical Biochemistry, 331, 283–295.
Jiye, A., Trygg, J., Gullberg, J., et al. (2005). Extraction and GC/MS analysis of the human blood plasma metabolome. Analytical Chemistry, 77, 8086–8094.
Jonsson, P., Johansson, E. S., Wuolikainen, A., et al. (2006). Predictive metabolite profiling applying hierarchical multivariate curve resolution to GC-MS Data-A potential tool for multi-parametric diagnosis. Journal of Proteome Research, 5, 1407–1414.
Kanani, H. H., & Klapa, M. I. (2007). Data correction strategy for metabolomics analysis using gas chromatography-mass spectrometry. Metabolic Engineering, 9, 39–51.
Kirkland, J. J. (1977). Sampling and extra-column effects in high-performance liquid chromatography; influence of peak skew on plate count calculations. Journal of Chromatographic Science, 15, 303–316.
Lapainis, T., Rubakhin, S. S., & Sweedler, J. V. (2009). Capillary electrophoresis with electrospray ionization mass spectrometric detection for single-cell metabolomics. Analytical Chemistry, 81, 5858–5864.
Pous-Torres, S., Baeza-Baeza, J. J., Torres-Lapasió, J. R., & García-Álvarez-Coque, M. C. (2008). Peak capacity estimation in isocratic elution. Journal of Chromatography A, 1205, 78–89.
Rodríguez, I., Quintana, J. B., Carpinteiro, J., Carro, A. M., Lorenzo, R. A., & Cela, R. (2003). Determination of acidic drugs in sewage water by gas chromatography-mass spectrometry as tert.-butyldimethylsilyl derivatives. Journal of Chromatography A, 985, 265–274.
Schweer, H. (1982). Gas chromatography–mass spectrometry of aldoses as O-methoxime, O-2-methyl-2-propoxime and O-n-butoxime pèrtrifluoroacetyl derivatives on OV-225 with methylpropane as ionization agent: I. Pentoses. Journal of Chromatography A, 236, 355–360.
Tam, Y. Y., & Normanly, J. (1998). Determination of indole-3-pyruvic acid levels in Arabidopsis thaliana by gas chromatography-selected ion monitoring-mass spectrometry. Journal of Chromatography A, 800, 101–108.
Wold, S. (1978). Cross-validatory estimation of the number of components in factor and principal components models. Technometrics, 20, 397–405.
Yu, Z., Peldszus, S., & Huck, P. M. (2007). Optimizing gas chromatographic-mass spectrometric analysis of selected pharmaceuticals and endocrine-disrupting substances in water using factorial experimental design. Journal of Chromatography A, 1148, 65–77.
Zelena, E., Dunn, W. B., Broadhurst, D., et al. (2009). Development of a robust and repeatable UPLC-MS method for the long-term metabolomic study of human serum. Analytical Chemistry, 81, 1357–1364.
Zhang, S., Nagana Gowda, G. A., Asiago, V., Shanaiah, N., Barbas, C., & Raftery, D. (2008). Correlative and quantitative 1H NMR-based metabolomics reveals specific metabolic pathway disturbances in diabetic rats. Analytical Biochemistry, 383, 76–84.
Acknowledgments
This work was supported by grants from Swedish Research Council (14196-06-3), the Crafoord Foundation, Lars Hierta, Fredrik and Ingrid Thuring, Åke Wiberg, Albert Påhlsson, O.E. and Edla Johansson Foundations, Knut and Alice Wallenberg Foundation, and the Royal Physiographic Society. Support from Inga and John Hain Foundation to PS is acknowledged.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Danielsson, A.P.H., Moritz, T., Mulder, H. et al. Development of a gas chromatography/mass spectrometry based metabolomics protocol by means of statistical experimental design. Metabolomics 8, 50–63 (2012). https://doi.org/10.1007/s11306-011-0283-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11306-011-0283-6