Abstract
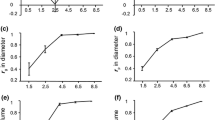
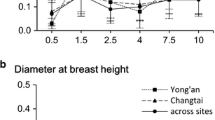
The objective of this study was to better understand the underlying gene action in eucalyptus, under different plantation densities, for a different set of traits: growth, bark thickness, ecophysiological, and wood chemical property traits. We estimated the magnitude and relative proportion of the various genetic variance components using a eucalyptus genotype by spacing (G × S) interaction experiment. A clonally replicated progeny test including 888 clones belonging to 64 full-sib families of Eucalyptus urophylla × Eucalyptus grandis hybrid was used to estimate genetic parameters using genomic information to assess relationship matrix. Two densities (833 and 2500 trees/ha) were used representing contrasted environments in terms of individual tree available resource. Results showed that for height and circumference, additive-by-spacing (A × S) interaction variance increased from 18 to 55 months old, while dominance-by-spacing (D × S) interaction variance decreased. For bark thickness, specific leaf area, nitrogen, calcium, and magnesium, A × S interaction variance was preponderant. For wood chemical properties, except with Klason lignin, genetic additive effects strongly interacted with spacing compared to non-additive effects.



Similar content being viewed by others
References
Akaike H (1974) A new look at the statistical model identification. Trans Autom Control 19:716–723
Bajgain P, Zhang X & Anderson JA (2019) Genome-wide association study of yield component traits in intermediate wheatgrass and implications in genomic selection and breeding. Genes Genomes Genetics: 9https://doi.org/10.1534/g3.119.400073
Bajgain P, Zhang X, Anderson JA (2020) Dominance and G×E interaction effects improve genomic prediction and genetic gain in intermediate wheatgrass (Thinopyrum intermedium). Plant Genome: e20012. https://doi.org/10.1002/tpg2.20012
Berlin M, Jansson G, Högberg K-A, Andreas Helmersson A (2019) Analysis of non –additive genetic effects in Norway spruce. Tree Genet Genomes 15:42. https://doi.org/10.1007/s11295-019-1350-9
Bijarpasi MM, Shahraji TR, Lahiji HS (2019) Genetic variability and heritability of some morphological and physiological traits in Fagus orientalis Lipsky along an elevation gradient in Hyrcanian forests. Folia Oecologica 46(1):45–53. https://doi.org/10.2478/foecol-2019-0007
Botrel MCG, Trugilho PF, Rosado SCdS, da Silva JRM (2010) Seleção de clones de Eucalyptus para biomassa florestal e qualidade da madeira. Scientia Forestalis, Picacicaba 38(86):237–245
Bouvet J-M, Makouanzi Ekomono CG, Brendel O, Laclau J-P, Bouillet J-P, Epron D (2020) Selecting for water use efficiency, wood chemical traits and biomass with genomic selection in a Eucalyptus breeding program. For Ecol Manage 465:118092. https://doi.org/10.1016/j.foreco.2020.118092
Bouvet J-M, Makouanzi G, Cros D and Vigneron Ph (2016) Modeling additive and non-additive effects in a hybrid population using genome-wide genotyping: prediction accuracy implications. Heredity: 1–12. https://doi.org/10.1038/hdy.2015.78
Bouvet J-M, Saya A, Vigneron Ph (2009) Trends in additive, dominance and environmental effects with age for growth traits in Eucalyptus hybrid populations. Euphytica 165:35–54
Bouvet J-M, Vigneron Ph (1996) Variance structure in Eucalyptus hybrid populations. Silvae Genetica 45(2–3):171–177
Bouvet J-M, Vigneron Ph, Gouma R, Saya A (2003) Trends in variances and heritabilities with age for growth traits in Eucalyptus spacing experiments. Silvae Genetica 52:121–133
Brito AS, Vidaurre GB, Oliveira JTdS, da Silva JGM, Oliveira RF, Júnior AFD, Arantes MDC, Moulin JC, Valin M, De Siqueira L, Zauza EAV (2021). Interaction between planting spacing and wood properties of Eucalyptus clones grown in short rotation. iForest 14 : 12–17. https://doi.org/10.3832/ifor3574-013
Burgueño J, Crossa J, Cornelius PL, Yang R-C (2008) Using factor analytic models for joining environments and genotypes without crossover genotype × environment interaction. Crop Sci. 48https://doi.org/10.2135/cropsci2007.11.0632
Burgueño J, de los Campos G, Weigel K and Crossa J, (2012) Genomic prediction of breeding values when modeling genotype × environment interaction using pedigree and dense molecular markers. Crop Sci 52:707–719
Calleja-Rodriguez A, Gull BA, Wu HX, Mullin TJ, Persson T (2019) Genotype-by-environment interactions and the dynamic relationship between tree vitality and height in northern Pinus sylvestris. Tree Genet Genomes 15:36. https://doi.org/10.1007/s11295-019-1343-8
Campbell RK, Echols RM, Stonecypher RW (1986) Genetic variances and interaction in 9-year-old Douglas fir grown at narrow spacings. Silvae Genetica 35:24–31
Chen Z-Q, Karlsson B, Wu HX (2017) Patterns of additive genotype-by-environment interaction in tree height of Norway spruce in southern and central Sweden. Tree Genet Genomes 13:25. https://doi.org/10.1007/s11295-017-1103-6
de La Torre AR, Wang T, Jaquish B, Aitken SN (2014) Adaptation and exogenous selection in a Picea glauca × Picea engelmannii hybrid zone: implications for forest management under climate change. New Phytol 201(2):687–699
de los Campos G, Sorensen DA, (2013) A commentary on Pitfalls of predicting complex traits from SNPs. Nat Rev Genet 14:894–894
Denis M, Favreau B, Ueno S, Camus-Kulandaivelu L, Chaix G, Gion JM, Nourrisier-Montou S, Polidori J, Bouvet J-M (2013) Genetic variation of wood chemical traits and association with underlying genes in Eucalyptus urophylla. Tree Genet Genomes 9:927–942
des Marais DL, Hernandez KM, Juenger TE (2013) Genotype-by environment interaction and plasticity: exploring genomic responses of plants to the abiotic environment. Annu Rev Ecol Evol Syst 44:5–29. https://doi.org/10.1146/annurev-ecolsys-110512-135806
Drost DR, Puranik S, Novaes E, Novaes CRDB, Dervinis C, Gailing O, Kirst M (2015) Genetical genomics of Populus leaf shape variation. BMC Plant Biol 15:1–10
Fox GP, Bowman J, Kelly A, Inkerman A, Poulsen D, Henry R (2007) Assessing for genetic and environmental effects on ruminant feed quality in barley (Hordeum vulgare). Euphytica 163:249–257. https://doi.org/10.1007/s10681-007-9638-5
Gammal El-Dien O, Ratcliffe B, Klápště J, Porth I, Chen C, El-Kassaby YA (2018) Multienvironment genomic variance decomposition analysis of open-pollinated Interior spruce (Picea glauca × engelmannii). Mol Breeding 38:26. https://doi.org/10.1007/s11032-018-0784-3
Gaspare WJ, Ivkovic´ M, Liepe K, Hamann A, Low CB, (2015) Drivers of genotype by environment interaction in radiata pine as indicated by multivariate regression trees. Forest Ecology Management 353:21–29
Gezan SA, de Carvalho MP, Sherrill J (2017) Statistical methods to explore genotype-by-environment interaction for loblolly pine clonal trials. Tree Genet Genomes 13:1–11. https://doi.org/10.1007/s11295-016-1081-0
Gilmour AR, Gogel BJ, Cullis BR and Thompson R (2006) ASREML user guide, Release 2.0. Hemel Hempstead, UK: VSN International, 342p.
Gouveia BT, Rios EF, Nunes JAR, Gezan SA, Munoz PR, Kenworthy KE, Unruh JB, Miller GL, Milla-Lewis SR, Schwartz BM, Raymer PL, Chandra A, WherleyBG WuY, Martin D, Moss JQ (2020) Genotype-by-environment interaction for turfgrass quality in bermudagrass across the southeastern United States. Crop Sci 60:3328–3343. https://doi.org/10.1002/csc2.20260
Gwaze DP, Wolliams JA, Kanowski PJ, Bridgwater FE (2001) Interactions of genotype with site for height and stem straightness in Pinus taeda in Zimbabwe. Silvae Genetica 50(3–4):135–140
Habier D, Fernando R, Dekkers J (2007) The impact of genetic relationship information on genome-assisted breeding values. Genetics 177(4):2389–2397
Hein PRG, Bouvet J-M, Mandrou E, Vigneron P, Clair B, Chaix G (2012) Age trends of microfibril angle inheritance and their genetic and environmental correlations with growth, density and chemical properties in Eucalyptus urophylla ST Blake wood. Ann For Sci 69(6):681–691. https://doi.org/10.1007/s13595-012-0186-3
Ignacio-Sánchez E, Vargas-Hernandez JJ, Lopez-Upton JY, Borja-de la Rosa A (2005) Parámetros genéticos del crecimiento y densidad de madera en edades juveniles de Eucalyptus urophylla s.t. Blake Agrociencia 39:469–479
Isaac-Renton M, Stoehr MU, Statland CB, Woods J (2020) Tree breeding and silviculture: Douglas-fir volume gains with minimal wood quality loss under variable planting densities. For Ecol Manage 465:118094. https://doi.org/10.1016/j.foreco.2020.118094
Kusnandar D, Galwer NW, Hertzler GL, Butcher TB (1998) Age trends in variances and heritabilities for diameter and height in maritime pine (Pinus pinaster Ait) in Western Australia. Silvae Genetica 47(2–3):136–141
Lai M, Dong L, Yi M, Sun S, Zhang Y, Fu L, Xu Z, Lei L, Leng C, Zhang L (2017) Genetic variation, heritability and genotype × environment interactions of resin yield, growth traits and morphologic traits for Pinus elliottii at Three Progeny Trials. Forests 8:409. https://doi.org/10.3390/f8110409
Li JH, Zhang QW, Su XH, Gao JS, Lu BM (2002) Multi-level genetic variation in leaf and growth of hybrid system between Populus deltoides and P. cathayana. Forest Res 15:76–82
Li Y, Suontama M, Burdon RD, Dungey HS (2017) Genotype by environment interaction in the forest tree breeding: review methodology and perspectives on research and application. Tree Genet Genomes 13:60. https://doi.org/10.1007/s11295-017-1144-x
Ling J, Xiao Y, Hu J, Wang F, Ouyang F, Wang J, Weng Y, Zhang H (2021) Genotype by environment interaction analysis of growth of Picea koraiensis families at diferent sites using BLUP-GGE. New Forest 52:113–127. https://doi.org/10.1007/s11056-020-09785-3
Ly D, Huet S, Gauffreteau A, Rincent R, Touzy G, Mini A, Jannink J-L, Cormier F, Paux E, Lafarge S, Le Gouis J, Charmet G (2018) Whole-genome prediction of reaction norms to environmental stress in bread wheat (Triticum aestivum L.) by genomic random regression. Field Crop Res 216:32–41
Makouanzi G, Chaix G, Nourissier S, Vigneron Ph (2018) Genetic variability of growth and wood chemical properties in a clonal population of Eucalyptus urophylla × Eucalyptus grandis in the Congo. South Forests: a Journal of Forest Science 80(2):151–158. https://doi.org/10.2989/20702620.2017.1298015
Mandrou E, Hein PRG, Villar E, Vigneron P, Plomion C, Gion J-M (2012) A candidate gene for lignin composition in Eucalyptus: cinnamoyl-CoA reductase (CCR). Tree Genet Genomes 8:353–364. https://doi.org/10.1007/s11295-011-0446-7
Mathews KL, Chapman SC, Trethowan R, PfeifferW GM, Crossa J, Payne T, DeLacy I, Fox PN, Cooper M (2007) Global adaptation patterns of Australian and CIMMYT spring bread wheat. Theor Appl Genet 115:819–835. https://doi.org/10.1007/s00122-007-0611-4
Oakey H, Cullis B, Thompson R, Comadran J, Halpin C & Robbie Waugh R (2016) Genomic selection in multi-environment crop trials. G3: https://doi.org/10.1534/g3.116.027524
Oehlert GW (1992) A note on the delta method. Am Statistician 46(1):27–29
Osorio LF, White TL, Huber DAH (2001) Age trends of heritabilities and genotype by-environment interactions for growth traits and wood density from clonal trials of Eucalyptus grandis HILL ex MAIDEN. Silvae Genetica 50(1):30–37
Patinot-Valera F & Kageyama PY (1995) Parametros geneticos y espaciamieto en progenie de Eucalyptus saligna Smith. Research Paper IPEF, Piracicaba (48/49): 61–76.
R Development Core Team (2011) R: a language and environment for statistical computing. Austria, R Foundation for Statistical Computing, Vienna
Rambolarimanana T, Ramamonjisoa L, Verhaegen D, Leong Pock Tsy J-M, Jacquin L, Cao-Hamadou T-V, Makouanzi G, Bouvet J-M (2018) Performance of multi-trait genomic selection for Eucalyptus robusta breeding program. Tree Genetics & Genomes 14:71. https://doi.org/10.1007/s11295-018-1286-5
Ren J, Ji X, Wang C, Hu J, Nervo G and Li J (2020) Variation and genetic parameters of leaf morphological traits of eight families from Populus simonii × P. nigra. Forests 11 (1319): 1–17. https://doi.org/10.3390/f11121319
Resende RT, Resende MDV, Silva FF, Azevedo CF, Takahashi EK, Silva-Junior OB, Grattapaglia D, D, (2017) Assessing the expected response to genomic selection of individuals and families in Eucalyptus breeding with an additive-dominant model. Heredity 1:11
Rönneberg-Wästjung AC, Gullberg U, Nilsson C (1994) Genetic parameters of growth characters in Salix viminalis grown in Sweden. Can J for Res 24:1060–1969
Russell DJF, Wanless S, Collingham YC, Huntley B, Hamer KC (2015) Predicting future European breeding distributions of British seabird species under climate change and unlimited/no dispersal scenarios. Diversity 7:342–359
Ryckewaert P, Chaix G, Héran D, Zgouz A, Bendoula R (2002) Evaluation of a combination of NIR micro-spectrometers to predict chemical properties of surgarcane forage using a multi-block approach. Biosys Eng 217:18–25. https://doi.org/10.1016/j.biosystemseng.2022.02.019
Sefton CA, Montagu K, Atwell BJ, Conroy JP (2002) Anatomical variation in juvenile eucalypt leaves accounts for differences in specific leaf area and CO2 assimilation rates. Austrian Journal of Botany 50:301–310
Smith AB, Ganesalingam A, Kuchel H, Cullis BR (2015) Factor analytic mixed models for the provision of grower information from national crop variety testing programs. Theor Appl Genet 128:55–72. https://doi.org/10.1007/s00122-014-2412-x
St Clair JB, Adams WT (1991) Relative family performance and variance structure of open pollinated Douglas-fir seedlings grown in three competitive environments. Theor Appl Genet 81:541–550
Stonecypher R, McCullough R (1981) Evaluation of full-sib families of Douglas fir in a nelder design. Proc South For Tree Improv Conf 16:56–76
Stuber CW, Cockerham CC (1966) Gene effects and variances in hybrid populations. Genetics 54:1279–1286
Souza CO, Silva JGM, Arantes MDC, Vidaurre GB, Dias Júnior AF, Oliveira MP (2020) Pyrolysis of Anadenanthera peregrina wood grown in different spacings from a forest plantation in Brazil aiming at the energy production. Environ Dev Sustain 22:5153–5168. https://doi.org/10.1007/s10668-019-00418-0
Tan B, Grattapaglia D, Wue HX, Ingvarsson PK (2018) Genomic relationships reveal significant dominance effects for growth in hybrid Eucalyptus. Plant Sci 267:84–93
Tanabe J, Endo R, Kuroda S, Ishiguri F, Narisawa T, Takashima Y (2019) Variance components and parent-offspring correlations of growth traits vary among the initial planting spacings in Zelkova serrata. Silvae Genetica 68:45–50
VanRaden PM (2008) Efficient methods to compute genomic predictions. J Dairy Sci 91(11):4414–4423
Volker PW, Potts BM, Borralho NMG (2008) Genetic parameters of intra- and inter-specific hybrids of Eucalyptus globulus and E. nitens. Tree Genet Genomes 4:445–460
Williams CG, Bridgwater FE, Lambeth CC (1983) Performance of single versus mixed family plantation blocks of loblolly pine. Proc South Forest Tree Improv Conf 17:56–76
Willman MR, Bushakra JM, Bassil N, Finn CE, Dossett M, Perkins-Veazie P, Bradish CM, Fernandez GE, Weber CA, Sheerens JC, Dunlap L, Fresnedo-Ramirez J (2022) Analysis of a multi- environment trial for black raspberry (Rubus occidentalis L.) quality traits. Genes (Basel) 13(3): 418. https://doi.org/10.3390/genes13030418
Xie CY, Ying CC (1996) Heritabilities, age-age correlations and early selection in lodgepole pine (Pinus contorta ssp. Latifolia). Sivae Genetica 45:101–107
Yang H, Weng Q, Li F, Zhou C, Li M, Chen S, Ji H & Gan S (2018) Genotypic variation and genotype-by-environment interactions in growth and wood properties in a cloned Eucalyptus urophylla × E. tereticornis family in Southern China. For Sci. 64 3 225 232
Acknowledgements
We are grateful to Philippe Vigneron for his contribution to the designed trial. We thank Crisley Ulrich Mayinguidi and all the technical team of CRDPI for maintaining the trial and for data collection. We would like also to thank Anne Clément-Vidal and Armelle Soutiras for their assistance with NIRS measurement, and Gilles Chaix for his assistance with the NIRS analysis. The authors thank the reviewers for their comments, which helped us improve the manuscript.
Author information
Authors and Affiliations
Contributions
CGME designed the trial. CGME and JMB supervised the collection of the field data. Near-infrared spectroscopy analyses were performed by CGME. CGME and JMB performed the statistical analyses and wrote the manuscript with the help of TR.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Communicated by V. Decroocq.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Makouanzi Ekomono, C.G., Rambolarimanana, T. & Bouvet, JM. Preponderance of additive and non-additive variances for growth, ecophysiological and wood traits in Eucalyptus hybrid genotype-by-spacing interaction. Tree Genetics & Genomes 18, 32 (2022). https://doi.org/10.1007/s11295-022-01563-w
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11295-022-01563-w