Abstract
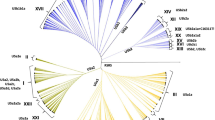
We aimed to test the extent to which plastid DNA gives incongruent phylogeographic patterns to nuclear DNA in a species of eucalypt, Eucalyptus behriana, a taxonomic group where chloroplast capture is a well-established phenomenon. Furthermore, we aimed to test the degree of influence chloroplast capture has on the observed patterns by broadly sampling co-occurring, related species. A genome skimming approach was used to sequence and assemble chloroplast genomes from population-level sampling of E. behriana, as well as samples of twenty-one other Eucalyptus section Adnataria species which co-occur with it. Phylogenetic analyses were first undertaken on just E. behriana to allow direct comparison to previously reported phylogeographic patterns based upon nuclear markers. A subsequent analysis including the related taxa was undertaken to investigate the degree of chloroplast capture and how this may be influencing the observed phylogeographic patterns. We found strong geographic structuring of plastid DNA relationships across the geographic range of E. behriana, with a basal divergence between the most northerly isolated population at West Wyalong and all other populations which does not match phylogeographic patterns based on nuclear markers. When outgroups were included, we found that E. behriana is highly polyphyletic with respect to all other species, starkly contrasting with the species well-supported monophylly based upon nuclear markers, and that chloroplast capture is so widespread that geographic patterns of the plastid genomes are consistent across species boundaries.




Similar content being viewed by others
Data availability
All data generated during this study are publicly available on the NCBI GenBank database and datasets used in analyses are included in the electronic supplementary material attached to this article.
References
Allal F, Sanou H, Millet L, Vaillant A, Camus-Kulandaivelu L, Logossa ZA, Lefèvre F, Bouvet JM (2011) Past climate changes explain the phylogeography of Vitellaria paradoxa over Africa. Heredity 107:174–186. https://doi.org/10.1038/hdy.2011.5
Alwadani KG, Janes JK, Andrew RL (2019) Chloroplast genome analysis of box-ironbark Eucalyptus. Mol Phylogenet Evol 136:76–86. https://doi.org/10.1016/j.ympev.2019.04.001
Atlas of Living Australia. (2019). Atlas of Living Australia. Retrieved 9 October 2019, from http://www.ala.org.au.
Australian Government. (2017). Australia’s bioregions (IBRA). from http://www.environment.gov.au/land/nrs/science/ibra.
Bayly MJ, Rigault P, Spokevicius A, Ladiges PY, Ades PK, Anderson C, Bossinger G, Merchant A, Udovicic F, Woodrow IE, Tibbits J (2013) Chloroplast genome analysis of Australian eucalypts – Eucalyptus, Corymbia, Angophora, Allosyncarpia and Stockwellia (Myrtaceae). Mol Phylogenet Evol 69:704–716. https://doi.org/10.1016/j.ympev.2013.07.006
Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL (2005) GenBank. Nucleic Acids Res 33:D34–D38. https://doi.org/10.1093/nar/gki063
Benson JS (2008) New South Wales vegetation classification and assessment. Part 2. Plant communities of the NSW South-western Slopes Bioregion and update of NSW Western Plains plant communities, Version 2 of the NSWVCA database. Cunninghamia 10:599–673
Booth TH (2017) Going nowhere fast: a review of seed dispersal in eucalypts. Aust J Bot 65:401–410. https://doi.org/10.1071/BT17019
Bowler J, Kotsonis A, Lawrence CR (2006) Environmental evolution of the Mallee region, western Murray Basin. Proc R Soc Victoria 118:161–210
Brooker MIH (2000) A new classification of the genus Eucalyptus L’Hér. (Myrtaceae). Aust Syst Bot 13:79–148. https://doi.org/10.1071/SB98008
Buzatti RSdO, Lemos-Filho JP, Bueno ML, Lovato MB (2017) Multiple Pleistocene refugia in the Brazilian cerrado: evidence from phylogeography and climatic nichemodelling of two Qualea species (Vochysiaceae). Bot J Linn Soc 185:307–320. https://doi.org/10.1093/botlinnean/box062
Connor DJ (1966) Vegetation studies in north-west Victoria I. the Beulah-Hopetoun area. Proc R Soc Victoria 79:579–595
Cook E (2009) A record of late Quaternary environments at lunette-lakes Bolac and Turangmoroke, Western Victoria, Australia, based on pollen and a range of non-pollen palynomorphs. Rev Palaeobot Palynol 153:185–224. https://doi.org/10.1016/j.revpalbo.2008.07.001
Council of Heads of Australasian Herbaria (2018) Australian Plant Census. IBIS database. Canberra, Centre for Australian National Biodiversity Research, Australian Government.
Crisp MD, Linder HP, Weston PH (1995) Cladistic biogeography of plants in Australia and New Guinea: congruent pattern reveals two endemic tropical tracks. Syst Biol 44:457–473. https://doi.org/10.2307/2413654
Currat M, Ruedi M, Petit RJ, Excoffier L (2008) The hidden side of invasions: massive introgression by local genes. Evolution 62:1908–1920. https://doi.org/10.1111/j.1558-5646.2008.00413.x
Dolman G, Joseph L (2015) Evolutionary history of birds across southern Australia: structure, history and taxonomic implications of mitochondrial DNA diversity in an ecologically diverse suite of species. Emu - Austral Ornithology 115:35–48. https://doi.org/10.1071/MU14047
Eaton DAR, Hipp A, González-Rodríguez A, Cavender-Bares J, Gonzalez Rodriguez A (2015) Historical introgression among the American live oaks and the comparative nature of tests for introgression. Evolution 69:2587–2601. https://doi.org/10.1111/evo.12758
Eaton DAR, Overcast I (2020) ipyrad: interactive assembly and analysis of RADseq datasets. Bioinformatics 36:2592–2594. https://doi.org/10.1093/bioinformatics/btz966
Fahey PS, Fowler RM, McLay TG, Udovicic F, Cantrill DJ, Bayly MJ (2021a) Divergent lineages in a semi-arid mallee species, Eucalyptus behriana, correspond to a major geographic break in south-eastern Australia. Ecol Evol 11:664–678. https://doi.org/10.1002/ece3.7099
Fahey PS, Nicolle D, McLay TGB, Udovicic F, Cantrill DJ, Bayly MJ (2021b) A phylogenetic investigation of the taxonomically problematic Eucalyptus odorata complex (Eucalyptus section Adnataria series Subbuxeales): evidence for extensive interspecific gene flow and reticulate evolution. Manuscript submitted for publication.
Flores-Rentería L, Rymer PD, Riegler M (2017) Unpacking boxes: integration of molecular, morphological and ecological approaches reveals extensive patterns of reticulate evolution in box eucalypts. Mol Phylogenet Evol 108:70–87. https://doi.org/10.1016/j.ympev.2017.01.019
French PA, Brown GK, Bayly MJ (2016) Incongruent patterns of nuclear and chloroplast variation in Correa (Rutaceae): introgression and biogeography in south-eastern Australia. Plant Syst Evol 302:447–468. https://doi.org/10.1007/s00606-016-1277-7
Gibbs A, Udovicic F, Drinnan A, Ladiges P (2009) Phylogeny and classification of Eucalyptus subgenus Eudesmia (Myrtaceae) based on nuclear ribosomal DNA, chloroplast DNA and morphology. Aust Syst Bot 22:158–179. https://doi.org/10.1071/SB08043
Griffin AR, Burgess IP, Wolf L (1988) Patterns of natural and manipulated hybridisation in the genus Eucalyptus L’Hérit - a review. Aust J Bot 36:41–66. https://doi.org/10.1071/BT9880041
Hayman D (1960) The distribution and cytology of the chromosome races of Themeda australis in southern Australia. Aust J Bot 8:58–68. https://doi.org/10.1071/BT9600058
Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS (2018) UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol 35:518–522. https://doi.org/10.1093/molbev/msx281
Huang D, Hefer C, Kolosova N, Douglas C, Cronk QCB (2014) Whole plastome sequencing reveals deep plastid divergence and cytonuclear discordance between closely related balsam poplars, Populus balsamifera and P. trichocarpa (Salicaceae). New Phytol 204:693–703. https://doi.org/10.1111/nph.12956
Jackson HD, Steane DA, Potts BM, Vaillancourt RE (1999) Chloroplast DNA evidence for reticulate evolution in Eucalyptus (Myrtaceae). Mol Ecol 8:739–751. https://doi.org/10.1046/j.1365-294X.1999.00614.x
Kalyaanamoorthy S, Minh BQ, Wong TK, von Haeseler A, Jermiin LS (2017) ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods 14:587–589. https://doi.org/10.1038/nmeth.4285
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. https://doi.org/10.1093/molbev/mst010
Kelly M, Mercer D (2005) Australia’s box–ironbark forests and woodlands: saving the fragments of a threatened ecosystem. Aust Geogr 36:19–37. https://doi.org/10.1080/00049180500050821
Kershaw AP, D’Costa DM, McEwen Mason JRC, Wagstaff BE (1991) Palynological evidence for Quaternary vegetation and environments of mainland southeastern Australia. Quatern Sci Rev 10:391–404. https://doi.org/10.1016/0277-3791(91)90003-D
Kershaw AP, McKenzie GM, Porch N, Roberts RG, Brown J, Heijnis H, Orr ML, Jacobsen G, Newall PR (2007) A high-resolution record of vegetation and climate through the last glacial cycle from Caledonia Fen, southeastern highlands of Australia. J Quat Sci 22:481–500. https://doi.org/10.1002/jqs.1127
Larcombe MJ, Holland B, Steane DA, Jones RC, Nicolle D, Vaillancourt RE, Potts BM (2015) Patterns of reproductive isolation in Eucalyptus—a phylogenetic perspective. Mol Biol Evol 32:1833–1846. https://doi.org/10.1093/molbev/msv063
Larcombe MJ, McKinnon GE, Vaillancourt RE (2011) Genetic evidence for the origins of range disjunctions in the Australian dry sclerophyll plant Hardenbergia violacea. J Biogeogr 38:125–136. https://doi.org/10.1111/j.1365-2699.2010.02391.x
Ley AC, Hardy OJ (2017) Hybridization and asymmetric introgression after secondary contact in two tropical African climber species, Haumania danckelmaniana and Haumania liebrechtsiana (Marantaceae). Int J Plant Sci 178:421–430. https://doi.org/10.1086/691628
Marginson JC, Ladiges PY (1988) Geographical variation in Eucalyptus baxteri s.l. and the recognition of a new species. E Arenacea Australian Systematic Botany 1:151–170. https://doi.org/10.1071/SB9880151
McKinnon GE, Vaillancourt R, Tilyard P, Potts B (2001) Maternal inheritance of the chloroplast genome in Eucalyptus globulus and interspecific hybrids. Genome 44:831–835. https://doi.org/10.1139/g01-078
McLay TGB. (2017). High quality DNA extraction protocol from recalcitrant plant tissues. from https://www.protocols.io/view/high-quality-dna-extraction-protocol-from-recalcit-i8jchun?more.
Myburg AA, Grattapaglia D, Tuskan GA, Hellsten U, Hayes RD, Grimwood J, Jenkins J, Lindquist E, Tice H, Bauer D, Goodstein DM, Dubchak I, Poliakov A, Mizrachi E, Kullan ARK, Hussey SG, Pinard D, van der Merwe K, Singh P, van Jaarsveld I, Silva-Junior OB, Togawa RC, Pappas MR, Faria DA, Sansaloni CP, Petroli CD, Yang X, Ranjan P, Tschaplinski TJ, Ye C-Y, Li T, Sterck L, Vanneste K, Murat F, Soler M, Clemente HS, Saidi N, Cassan-Wang H, Dunand C, Hefer CA, Bornberg-Bauer E, Kersting AR, Vining K, Amarasinghe V, Ranik M, Naithani S, Elser J, Boyd AE, Liston A, Spatafora JW, Dharmwardhana P, Raja R, Sullivan C, Romanel E, Alves-Ferreira M, Külheim C, Foley W, Carocha V, Paiva J, Kudrna D, Brommonschenkel SH, Pasquali G, Byrne M, Rigault P, Tibbits J, Spokevicius A, Jones RC, Steane DA, Vaillancourt RE, Potts BM, Joubert F, Barry K, Pappas GJ, Strauss SH, Jaiswal P, Grima-Pettenati J, Salse J, Van de Peer Y, Rokhsar DS, Schmutz J (2014) The genome of Eucalyptus grandis. Nature 510:356–362. https://doi.org/10.1038/nature13308
Nevill PG, Després T, Bayly MJ, Bossinger G, Ades PK (2014) Shared phylogeographic patterns and widespread chloroplast haplotype sharing in Eucalyptus species with different ecological tolerances. Tree Genet Genomes 10:1079–1092. https://doi.org/10.1007/s11295-014-0744-y
Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ (2015) IQ-TREE: a fast and effective stochastic algorithm for estimating maximum likelihood phylogenies. Molecular Biology and Evolution 32:268–274. https://doi.org/10.1093/molbev/msu300
Nicolle D (2014) Myrtaceae (partly) (version 1).In: J. Kellermann (ed.) Flora of South Australia. (5th edition).102 pp. State Herbarium of South Australia, Adelaide.
Nicolle D. (2019). Classification of the eucalypts (Angophora, Corymbia and Eucalyptus) version 4. from https://www.dn.com.au/Classification-Of-The-Eucalypts.pdf.
Owens GL, Baute GJ, Rieseberg LH (2016) Revisiting a classic case of introgression: hybridization and gene flow in Californian sunflowers. Mol Ecol 25:2630–2643. https://doi.org/10.1111/mec.13569
Paradis E (2010) pegas: an R package for population genetics with an integrated–modular approach. Bioinformatics 26:419–420. https://doi.org/10.1093/bioinformatics/btp696
Payn K, Dvorak W, Myburg A (2007) Chloroplast DNA phylogeography reveals the island colonisation route of Eucalyptus urophylla (Myrtaceae). Aust J Bot 55:673–683. https://doi.org/10.1071/BT07056
Petit RJ, Csaikl UM, Bordacs S, Burg K, Coart E, Cottrell J, van Dam B, Deans JD, Dumolin-Lapegue S, Fineschi S, Finkeldey R, Gillies A, Glaz I, Goicoechea PG, Jensen JS, Konig AO, Lowe AJ, Madsen SF, Matyas G, Munro RC, Olalde M, Pemonge MH, Popescu F, Slade D, Tabbener H, Taurchini D, de Vries SGM, Ziegenhagen B, Kremer A (2002) Chloroplast DNA variation in European white oaks - phylogeography and patterns of diversity based on data from over 2600 populations. For Ecol Manage 156:5–26. https://doi.org/10.1016/s0378-1127(01)00645-4
Pollock L, Bayly M, Nevill P, Vesk P (2013) Chloroplast DNA diversity associated with protected slopes and valleys for hybridizing Eucalyptus species on isolated ranges in south-eastern Australia. J Biogeogr 40:155–167. https://doi.org/10.1111/j.1365-2699.2012.02766.x
Rieseberg LH, Soltis DE (1991) Phylogenetic consequences of cytoplasmic gene flow in plants. Evolutionary Trends in Plants 5:65–84
Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61:539–542. https://doi.org/10.1093/sysbio/sys029
Schuster TM, Setaro SD, Tibbits JFG, Batty EL, Fowler R, McLay TGB, Wilcox S, Ades PK, Bayly MJ (2018) Chloroplast variation is incongruent with classification of the Australian bloodwood eucalypts (genus Corymbia, family Myrtaceae). PLoS ONE 13:e0195034. https://doi.org/10.1371/journal.pone.0195034
Slee AV, Brooker MIH, Duffy S, West J (2015) EUCLID: Eucalypts of Australia. 4th ed. (internet based, hosted by the Identic Pty Ltd, Brisbane.) Centre for Australian National Biodiversity Research, Canberra.
Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. https://doi.org/10.1093/bioinformatics/btu033
Steane D, Byrne M, Vaillancourt RE, Potts BM (1998) Chloroplast DNA polymorphism signals complex interspecific interactions in Eucalyptus (Myrtaceae). Aust Syst Bot 11:25–40. https://doi.org/10.1071/SB96028
Swofford D (2002) PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods). 4th ed. Sinauer Associates, Sunderland, Massachusetts.
Thornhill AH, Crisp MD, Külheim C, Lam KE, Nelson LA, Yeates DK, Miller JT (2019) A dated molecular perspective of eucalypt taxonomy, evolution and diversification. Aust Syst Bot 32:29–48. https://doi.org/10.1071/SB18015
Zhang Q, Chiang TY, George M, Liu JQ, Abbott RJ (2005) Phylogeography of the Qinghai-Tibetan Plateau endemic Juniperus przewalskii (Cupressaceae) inferred from chloroplast DNA sequence variation. Mol Ecol 14:3513–3524. https://doi.org/10.1111/j.1365-294X.2005.02677.x
Acknowledgements
We thank Stephen Wilcox of the Walter and Eliza Hall Institute for assistance with DNA sequencing. Patrick Fahey is supported by the Jim Ross PhD Studentship from the Cybec Foundation and the Royal Botanic Gardens Victoria and is grateful for additional support from The University of Melbourne Botany Foundation. Collections for this study were completed under permits granted by the Department of Environment, Land, Water and Planning in Victoria (10008557), NSW National Parks & Wildlife Service (SL102100) and Department for Environment and Water in South Australia (Q26766).
Funding
Research costs were supported by a grant from Eucalypt Australia (grant number 2016–42).
Author information
Authors and Affiliations
Contributions
Patrick Fahey, Frank Udovicic, David Cantrill and Michael Bayly conceived the ideas; Patrick Fahey, Frank Udovicic, David Cantrill, Rachael Fowler and Michael Bayly conducted the fieldwork; Patrick Fahey and Rachael Fowler generated and analysed the data and Patrick Fahey led the writing with assistance from Frank Udovicic, David Cantrill, Rachael Fowler and Michael Bayly.
Corresponding author
Ethics declarations
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest
The authors declare no competing interests.
Additional information
Communicated by J.P. Jaramillo-Correa
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Fahey, P.S., Fowler, R.M., Udovicic, F. et al. Use of plastid genome sequences in phylogeographic studies of tree species can be misleading without comprehensive sampling of co-occurring, related species. Tree Genetics & Genomes 17, 43 (2021). https://doi.org/10.1007/s11295-021-01524-9
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11295-021-01524-9