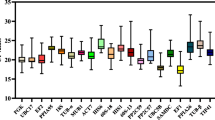
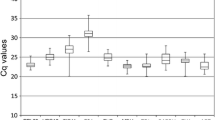
Abstract
Quantitative real-time PCR (RT-qPCR) techniques have revolutionized gene expression analyses. To obtain accurate results, raw RT-qPCR results need to be normalized by using endogenous reference genes whose expression is assumed invariable in all studied samples. However, there are no universal reference genes, and candidate genes need to be evaluated for each experimental condition. In this work, we tested a set of possible reference genes for use in different organs and tissues of Pinus pinaster (needles from adult trees and different organs and developmental stages of seedlings). The putative reference genes were selected using microarray analyses and from those commonly used in previous works. To achieve reproducible and reliable results, Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) guidelines were followed. To highlight the importance of these rules, 10 alternative primer pairs to be evaluated in pine samples were designed by following or not following the MIQE guidelines. Twenty-four candidate reference genes were tested in pine needles and 14 were also tested in pine seedlings. In both cases, valid reference genes were found, but differences in the stability and expression levels were also observed. Furthermore, a few of the best genes had unknown functions. The five most stable genes in the pine seedlings as well as four new candidate reference genes were evaluated in isolated tissues using laser capture microdissection. The results showed that the appropriate reference genes in different maritime pine organs were not invariable when sourced from the different tissues forming the organs.







Similar content being viewed by others
References
Andersen CL, Jensen JL, Orntoft TF (2004) Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 64:5245–5250. doi:10.1158/0008-5472.CAN-04-0496
Barber RD, Harmer DW, Coleman RA, Clark BJ (2005) GAPDH as a housekeeping gene: analysis of GAPDH mRNA expression in a panel of 72 human tissues. Physiol Genomics 21:389–395. doi:10.1152/physiolgenomics.00025.2005
Barsalobres-Cavallari CF, Severino FE, Maluf MP, Maia IG (2009) Identification of suitable internal control genes for expression studies in Coffea arabica under different experimental conditions. BMC Mol Biol 10:1. doi:10.1186/1471-2199-10-1
Boggy GJ, Woolf PJ (2010) A mechanistic model of PCR for accurate quantification of quantitative PCR data. PLoS One 5:e12355. doi:10.1371/journal.pone.0012355
Brunner AM, Yakovlev IA, Strauss SH (2004) Validating internal controls for quantitative plant gene expression studies. BMC Plant Biol 4:14. doi:10.1186/1471-2229-4-14
Bustin SA, Benes V, Nolan T, Pfaffl MW (2005) Quantitative real-time RT-PCR—a perspective. J Mol Endocrinol 34:597–601. doi:10.1677/jme.1.01755
Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT (2009) The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55:611–622. doi:10.1373/clinchem.2008.112797
Bustin SA, Beaulieu JF, Huggett J, Jaggi R, Kibenge FS, Olsvik PA, Penning LC, Toegel S (2010) MIQE precis: practical implementation of minimum standard guidelines for fluorescence-based quantitative real-time PCR experiments. BMC Mol Biol 11:74. doi:10.1186/1471-2199-11-74
Bustin SA, Benes V, Garson J, Hellemans J, Huggett J, Kubista M et al (2013) The need for transparency and good practices in the RT-qPCR literature. Nat Methods 10:1063–1067. doi:10.1038/nmeth.2697
Canales J, Flores-Monterrosso A, Rueda-Lopez M, Avila C, Canovas FM (2010) Identification of genes regulated by ammonium availability in the roots of maritime pine trees. Amino Acids 39:991–1001. doi:10.1007/s00726-010-0483-9
Canales J, Rueda-Lopez M, Craven-Bartle B, Avila C, Canovas FM (2012) Novel insights into regulation of asparagine synthetase in conifers. Front Plant Sci 3:100. doi:10.3389/fpls.2012.00100
Canales J, Bautista R, Label P, Gomez-Maldonado J, Lesur I, Fernandez-Pozo N, Rueda-Lopez M, Guerrero-Fernandez D, Castro-Rodriguez V, Benzekri H, Cañas RA, Guevara MA, Rodrigues A, Seoane P, Teyssier C, Morel A, Ehrenmann F, Le Provost G, Lalanne C, Noirot C, Klopp C, Reymond I, Garcia-Gutierrez A, Trontin JF, Lelu-Walter MA, Miguel C, Cervera MT, Canton FR, Plomion C, Harvengt L, Avila C, Gonzalo Claros M, Canovas FM (2014) De novo assembly of maritime pine transcriptome: implications for forest breeding and biotechnology. Plant Biotech J 12:286–299. doi:10.1111/pbi.12136
Cañas RA, de la Torre F, Canovas FM, Canton FR (2007) Coordination of PsAS1 and PsASPG expression controls timing of re-allocated N utilization in hypocotyls of pine seedlings. Planta 225:1205–1219. doi:10.1007/s00425-006-0431-9
Cañas RA, Canales J, Gomez-Maldonado J, Avila C, Canovas FM (2014) Transcriptome analysis in maritime pine using laser capture microdissection and 454 pyrosequencing. Tree Physiol 34:1278–1288. doi:10.1093/treephys/tpt113
Cañas RA, Canales J, Munoz-Hernandez C, Granados JM, Avila C, Garcia-Martin ML, Canovas FM (2015a) Understanding developmental and adaptive cues in pine through metabolite profiling and co-expression network analysis. J Exp Bot 66:3113–3127. doi:10.1093/jxb/erv118
Cañas RA, Feito I, Fuente-Maqueda JF, Ávila C, Majada J, Cánovas FM (2015b) Transcriptome-wide analysis supports environmental adaptations of two Pinus pinaster populations from contrasting habitats. BMC Genomics 16:909. doi:10.1186/s12864-015-2177-x
Chang E, Shi S, Liu J, Cheng T, Xue L, Yang X, Yang W, Lan Q, Jiang Z (2012) Selection of reference genes for quantitative gene expression studies in Platycladus orientalis (Cupressaceae) Using real-time PCR. PLoS One 7:e33278. doi:10.1371/journal.pone.0033278
Corpet F (1988) Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res 16:10881–10890. doi:10.1093/nar/16.22.10881
de Oliveira LA, Breton MC, Bastolla FM, Camargo Sda S, Margis R, Frazzon J, Pasquali G (2012) Reference genes for the normalization of gene expression in eucalyptus species. Plant Cell Physiol 53:405–422. doi:10.1093/pcp/pcr187
de Vega-Bartol JJ, Santos RR, Simoes M, Miguel CM (2013) Normalizing gene expression by quantitative PCR during somatic embryogenesis in two representative conifer species: Pinus pinaster and Picea abies. Plant Cell Rep 32:715–729. doi:10.1007/s00299-013-1407-4
Derveaux S, Vandesompele J, Hellemans J (2010) How to do successful gene expression analysis using real-time PCR. Methods 50:227–230. doi:10.1016/j.ymeth.2009.11.001
Dheda K, Huggett JF, Bustin SA, Johnson MA, Rook G, Zumla A (2004) Validation of housekeeping genes for normalizing RNA expression in real-time PCR. BioTechniques 37:112–114. doi:10.2144/3701A0112
Dheda K, Huggett JF, Chang JS, Kim LU, Bustin SA, Johnson MA, Rook GA, Zumla A (2005) The implications of using an inappropriate reference gene for real-time reverse transcription PCR data normalization. Anal Biochem 344:141–143. doi:10.1016/j.ab.2005.05.022
Farjon A (2010) A handbook of the world’s conifers. Brill, Leiden, Boston. doi:10.1163/9789047430629
Fink T, Lund P, Pilgaard L, Rasmussen JG, Duroux M, Zachar V (2008) Instability of standard PCR reference genes in adipose-derived stem cells during propagation, differentiation and hypoxic exposure. BMC Mol Biol 9:98. doi:10.1186/1471-2199-9-98
Fleige S, Pfaffl MW (2006) RNA integrity and the effect on the real-time qRT-PCR performance. Mol Aspects Med 27:126–139. doi:10.1016/j.mam.2005.12.003
Gibson UE, Heid CA, Williams PM (1996) A novel method for real time quantitative RT-PCR. Genome Res 6:995–1001. doi:10.1101/gr.6.10.995
Guénin S, Mauriat M, Pelloux J, Van Wuytswinkel O, Bellini C, Gutierrez L (2009) Normalization of qRT-PCR data: the necessity of adopting a systematic, experimental conditions-specific, validation of references. J Exp Bot 60:487–493. doi:10.1093/jxb/ern305
Gutierrez L, Mauriat M, Guenin S, Pelloux J, Lefebvre JF, Louvet R, Rusterucci C, Moritz T, Guerineau F, Bellini C, Van Wuytswinkel O (2008) The lack of a systematic validation of reference genes: a serious pitfall undervalued in reverse transcription-polymerase chain reaction (RT-PCR) analysis in plants. Plant Biotech J 6:609–618. doi:10.1111/j.1467-7652.2008.00346.x
Heid CA, Stevens J, Livak KJ, Williams PM (1996) Real time quantitative PCR. Genome Res 6:986–994. doi:10.1101/gr.6.10.986
Hellemans J, Vandesompele J (2014) Selection of reliable reference genes for RT-RT-qPCR analysis. Method Mol Biol 1160:19–26. doi:10.1007/978-1-4939-0733-5_3
Hruz T, Wyss M, Docquier M, Pfaffl MW, Masanetz S, Borghi L, Verbrugghe P, Kalaydjieva L, Bleuler S, Laule O, Descombes P, Gruissem W, Zimmermann P (2011) RefGenes: identification of reliable and condition specific reference genes for RT-RT-qPCR data normalization. BMC Genomics 12:156. doi:10.1186/1471-2164-12-156
Huang T, Bohlenius H, Eriksson S, Parcy F, Nilsson O (2005) The mRNA of the Arabidopsis gene FT moves from leaf to shoot apex and induces flowering. Science 309:1694–1696. doi:10.1126/science.1117768
Huggett J, Dheda K, Bustin S, Zumla A (2005) Real-time RT-PCR normalisation; strategies and considerations. Genes Immun 6:279–284. doi:10.1038/sj.gene.6364190
Kozera B, Rapacz M (2013) Reference genes in real-time PCR. J Appl Genet 54:391–406. doi:10.1007/s13353-013-0173-x
Li L, Ji G, Ye C, Shu C, Zhang J, Liang C (2015) PlantOrDB: a genome-wide ortholog database for land plants and green algae. BMC Plant Biol 15:161. doi:10.1186/s12870-015-0531-4
Liu D, Shi L, Han C, Yu J, Li D, Zhang Y (2012) Validation of reference genes for gene expression studies in virus-infected Nicotiana benthamiana using quantitative real-time PCR. PLoS One 7:e46451. doi:10.1371/journal.pone.0046451
Lovdal T, Lillo C (2009) Reference gene selection for quantitative real-time PCR normalization in tomato subjected to nitrogen, cold, and light stress. Anal Biochem 387:238–242. doi:10.1016/j.ab.2009.01.024
Mallona I, Lischewski S, Weiss J, Hause B, Egea-Cortines M (2010) Validation of reference genes for quantitative real-time PCR during leaf and flower development in Petunia hybrida. BMC Plant Biol 10:4. doi:10.1186/1471-2229-10-4
Marum L, Miguel A, Ricardo CP, Miguel C (2012) Reference gene selection for quantitative real-time PCR normalization in Quercus suber. PLoS One 7:e35113. doi:10.1371/journal.pone.0035113
Mehdi Khanlou K, Van Bockstaele E (2012) A critique of widely used normalization software tools and an alternative method to identify reliable reference genes in red clover (Trifolium pratense L.). Planta 236:1381–1393. doi:10.1007/s00425-012-1682-2
Meyer K, Koester T, Staiger D (2015) Pre-mRNA splicing in plants: in vivo functions of RNA-binding proteins implicated in the splicing process. Biogeosciences 5:1717–1740. doi:10.3390/biom5031717
Michaeli S, Galili G (2014) Degradation of organelles or specific organelle components via selective autophagy in plant cells. Int J Mol Sci 15:7624–7638. doi:10.3390/ijms15057624
Mullis KB, Faloona FA (1987) Specific synthesis of DNA in vitro via a polymerase-catalyzed chain reaction. Method Enzymol 155:335–350. doi:10.1016/0076-6879(87)55023-6
Oakley RV, Wang YS, Ramakrishna W, Harding SA, Tsai CJ (2007) Differential expansion and expression of alpha- and beta-tubulin gene families in Populus. Plant Pphysiol 145:961–973. doi:10.1104/pp.107.107086
Pfaffl M, Hageleit M (2001) Validities of mRNA quantification using recombinant RNA and recombinant DNA external calibration curves in real-time RT-PCR. Biotechnol Lett 23:275–282. doi:10.1023/A:1005658330108
Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP (2004) Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper—Excel-based tool using pair-wise correlations. Biotechnol Lett 26:509–515. doi:10.1023/B:BILE.0000019559.84305.47
Ruijter JM, Ramakers C, Hoogaars WM, Karlen Y, Bakker O, van den Hoff MJ, Moorman AF (2009) Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res 37:e45. doi:10.1093/nar/gkp045
Saha P, Blumwald E (2014) Assessing reference genes for accurate transcript normalization using quantitative real-time PCR in pearl millet [Pennisetum glaucum (L.) R. Br]. PLoS One 9:e106308. doi:10.1371/journal.pone.0106308
Spanakis E (1993) Problems related to the interpretation of autoradiographic data on gene expression using common constitutive transcripts as controls. Nucleic Acids Res 21:3809–3819. doi:10.1093/nar/21.16.3809
Suzuki T, Higgins PJ, Crawford DR (2000) Control selection for RNA quantitation. BioTechniques 29:332–337
Taylor S, Wakem M, Dijkman G, Alsarraj M, Nguyen M (2010) A practical approach to RT-RT-qPCR-Publishing data that conform to the MIQE guidelines. Methods 50:S1–S5. doi:10.1016/j.ymeth.2010.01.005
Theissen G (2002) Secret life of genes. Nature 415:741. doi:10.1038/415741a
Thellin O, Zorzi W, Lakaye B, De Borman B, Coumans B, Hennen G, Grisar T, Igout A, Heinen E (1999) Housekeeping genes as internal standards: use and limits. J Biotechnol 75:291–295. doi:10.1016/S0168-1656(99)00163-7
Tusher VG, Tibshirani R, Chu G (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A 98:5116–5121. doi:10.1073/pnas.091062498
Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG (2012) Primer3—new capabilities and interfaces. Nucleic Acids Res 40:e115. doi:10.1093/nar/gks596
Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034
Vestman D, Larsson E, Uddenberg D, Cairney J, Clapham D, von Arnold S (2011) Important processes during differentiation and early development of somatic embryos of Norway spruce as revealed by changes in global gene expression. Tree Genet Genomes 7:347–362. doi:10.1007/s11295-010-0336-4
Wang HL, Chen J, Tian Q, Wang S, Xia X, Yin W (2014) Identification and validation of reference genes for Populus euphratica gene expression analysis during abiotic stresses by quantitative real-time PCR. Physiol Plantarum 152:529–545. doi:10.1111/ppl.12206
Wei L, Miao H, Zhao R, Han X, Zhang T, Zhang H (2013) Identification and testing of reference genes for Sesame gene expression analysis by quantitative real-time PCR. Planta 237:873–889. doi:10.1007/s00425-012-1805-9
Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL (2012) Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bbioinformatics 13:134. doi:10.1186/1471-2105-13-134
Zimmermann P, Hirsch Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Pphysiol 136:2621–2632. doi:10.1104/pp.104.046367
Zuker M (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31:3406–3415. doi:10.1093/nar/gkg595
Acknowledgments
This work was supported by the European Commission Seventh Framework grant PROCOGEN (FP7-KBBE-2011-5), the Ministerio de Economía y Competitividad grant (BIO2015-69285-R), and Junta de Andalucía (BIO2012-0474).
Data archiving statement
Primer sequences are provided in Tables 1 and 2. Transcripts sequences are deposited in SustainPineDB (http://www.scbi.uma.es/sustainpinedb/home_page). Microarray data are available in NCBI GEO database (GSE55868).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by P. Ingvarsson
Electronic Supplementary Materials
Below is the link to the electronic supplementary material.
ESM 1
Supplemental Data S1. Cq values for the primer efficiency calculation. Four different calculations were included (PDF 124 kb)
ESM 2
Supplemental Data S2. Cq values for MIQE and Non-MIQE compliant primers in the all adult needle samples. The table includes the stability index calculated in geNorm (PDF 209 kb)
ESM 3
Supplemental Data S3. Cq values for MIQE compliant primers in the adult needle samples. The table includes the stability index calculated in geNorm, NormFinder, and BestKeeper (PDF 189 kb)
ESM 4
Supplemental Data S4. Cq values for MIQE compliant primers in the embryo and seedling samples. The table includes the stability index calculated in geNorm, Normfinder and Bestkeeper. (PDF 190 kb)
ESM 5
Supplemental Data S5. Cq values for the three better candidate reference genes in different physiological conditions and tissues. The table includes the stability index calculated in geNorm, NormFinder, and BestKeeper (PDF 208 kb)
ESM 6
Supplemental Data S6. Cq values for MIQE compliant primers at Laser Capture Microdissection samples. The table includes the stability index calculated in geNorm, NormFinder, and BestKeeper (PDF 192 kb)
Rights and permissions
About this article
Cite this article
Granados, J.M., Ávila, C., Cánovas, F.M. et al. Selection and testing of reference genes for accurate RT-qPCR in adult needles and seedlings of maritime pine. Tree Genetics & Genomes 12, 60 (2016). https://doi.org/10.1007/s11295-016-1018-7
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11295-016-1018-7