Abstract
A molecular marker analysis for Asian persimmon cultivar relationships and pollination status was conducted with 496 amplified fragment length polymorphism (AFLP) markers and 146 cultivars of Asian origin. Cultivars from China, Korea, and Japan were evaluated for marker composition and pollination status, which strongly influences fruit characteristics. Separation of Chinese, Korean, and Japanese cultivar groups and pollination type by neighbor-joining clustering, multidimensional scaling, and STRUCTURE was only weakly supported and not genetically significant. Significant differences for cultivar origin and pollination status were found for analysis of molecular variance (AMOVA), but most of the variation was among cultivars, not classification groups. All of the cultivar groups were genetically similar at the molecular level with most polymorphism due to individual cultivar differences.
Similar content being viewed by others
Introduction
Persimmon (Diospyros kaki Thunb.) is one of the most important fruit crops in East Asia and the fourth most important Japanese fruit. In 2012, 254,000 t of persimmon was produced in Japan (Japan Statistical Yearbook 2015). Diospyros kaki is an autohexaploid species (Tamura et al. 1998). Persimmon cultivars are classified into four types depending on the nature of astringency loss in the fruit and change in flesh color (Hume 1913, 1914; Kajiura 1946; Yonemori et al. 2000). Classifications are pollination-constant non-astringent (PCNA), pollination-variant non-astringent (PVNA), pollination-variant astringent (PVA), and pollination-constant astringent (PCA). Pollination constant cultivars retain their original flesh color, while the flesh of pollination variant cultivars becomes darker. Among these four types, the PCNA and PVNA types lose astringency naturally during fruit growth and become edible at maturity, while PVA and PCA types retain their astringency until fruit softening. Non-astringent cultivars are desired since they do not require postharvest treatment to remove astringency. PVNA cultivars require pollination and seed formation to lose astringency associated with flesh browning. PCNA cultivars are highly desired because they lose their astringency on the tree without needing complete pollination or seed formation. Persimmons probably originated in China and have been grown in China, Korea, and Japan as an important food source from prehistoric times. Many cultivars have been developed in these three countries during a long history of cultivation. Non-astringent-type cultivars, both PVNA and PCNA cultivars, were developed in Japan and China (Du et al. 2009; Yonemori et al. 2005). According to historical records in Japan, the oldest PVNA-type cultivar ‘Zenjimaru’ was found in 1214 ad and ‘Gosho,’ assumed to be the first PCNA-type cultivar, was first documented in the 17th century (Kikuchi 1948). A nationwide survey of persimmon cultivars in Japan (Agricultural Research Station 1912) identified only six PCNA-type cultivars vs. 401 PVNA-type cultivars among more than 1000 cultivars collected throughout Japan. The origin of PCNA cultivars is limited to the central part of Japan, while PVNA cultivars were developed throughout Japan (Yonemori et al. 2000). PCNA cultivars probably have more recent origins and very limited genetic variability. There are 963 persimmon cultivars reported in China (Renzi et al. 1997) and 186 cultivars described in Korea (Cho and Cho 1965). Most of these cultivars are reported to be astringent-type. One named cultivar, ‘Luo Tian Tian Shi,’ and several other genotypes have been reported from Luotian County in Hubei province of China as PCNA types (Yonemori et al. 2005).
Astringency in persimmon fruits is caused by tannins, which accumulate in vacuoles of idioblasts from flesh parenchyma cells (tannin cells). Production of ethanol and acetaldehyde by the seeds is associated with the loss of astringency in PVNA-type fruits but not in PCNA-type fruits. The seeds of PVNA-type fruits produce a large amount of ethanol and acetaldehyde during the middle stages of fruit development. These volatile compounds, especially acetaldehyde, cause coagulation of tannins in the large tannin cells, which results in the complete loss of astringency. After completion of tannin coagulation, tannin cells become brown through further oxidative reactions, producing a dark-colored flesh in PVNA-type fruits. The seeds of PVA-type fruits also produce these volatile compounds during the fruit development, but in limited amounts, so that the coagulation of tannins is restricted around the seeds and astringency remains in the rest of the flesh. Dark flesh color caused by tannin coagulation is also restricted around the seeds in PVA-type fruits. The seeds of PCA-type fruits produce almost no ethanol and acetaldehyde during their development; therefore, PCA-type fruits do not lose astringency naturally on the tree. The ability of seeds to generate these volatile compounds is high in the PVNA type, low in the PCA type, and intermediate in the PVA-type fruits, suggesting that the appearance of the PVNA character is due to a constitutive change in the ability of the fruit to produce ethanol and acetaldehyde in the seeds. This change appears to be a quantitative trait. ‘Zenjimaru,’ the first PVNA cultivar, probably appeared as the result of mutations from the PCA cultivar type to produce ethanol and acetaldehyde in the seed.
PCNA-type cultivars appear to have originated from a mutation causing loss of the ability to accumulate tannins in the fruit at an early stage of fruit development; this is a qualitative trait (Yonemori et al. 2000; Akagi et al. 2009, 2010, 2012). The first PCNA-type cultivar is thought to have been derived from PCA cultivars (Yonemori et al. 2000). The mutation probably occurred much more recently than the PCA to PVNA seed mutations. In addition to the PCNA cultivars in Japan, the Chinese PCNA cultivar, ‘Luo Tian Tian Shi,’ also loses the ability to accumulate tannins at an early stage of fruit development. However, the genetic basis for this PCNA trait is different than for the Japanese cultivars. It appears to be a dominant mutation (Ikegami et al. 2006, 2011) rather than the recessive trait found in the Japanese cultivars (Akagi et al. 2009, 2010, 2012; Kanzaki et al. 2001).
Several publications describe genetic diversity in European persimmon and relationships with Japanese cultivars (Badenes et al. 2003; Yonemori et al. 2008a, b; Naval et al. 2010). Diversity in Japanese and Chinese persimmons has been studied by Guo and Luo (2006, 2011) using SRAPs and simple sequence repeats (SSRs) and Yamagishi et al. (2005) with RAPDs. Kanzaki et al. (2000) studied the relationships among 19 Japanese PCNA and 14 non-PCNA cultivars with 138 amplified fragment length polymorphisms (AFLPs) for comparison with the Chinese PCNA cultivar, ‘Luo Tian Tian Shi.’ We evaluated large populations of cultivars developed in different parts of Asia, with the four types of fruit development and representing the complete range of cultivar diversity found in China, Japan, and Korea in the present study. The analysis of Asian cultivar relationships by Yonemori et al. (2008b) was extended using improved analytical methods. Genetic relationships among 146 Asian persimmon cultivars from China, Japan, and Korea were evaluated with 496 AFLP molecular makers in this study.
Materials and methods
Plant materials
Young leaves were collected from 153 persimmon cultivars at the Department of Grape and Persimmon Research Station, National Institute of Fruit Tree, or at the Experimental Farm of the Graduate School of Agriculture, Kyoto University germplasm collection. Seven cultivars were not used due to nomenclature and pollination status questions. Of the 146 cultivars analyzed, 102 cultivars are native to Japan, including 33 PCNA-, 27 PVNA-, 9 PVA-, and 33 PCA-type cultivars, 20 cultivars are native to Korea, and 24 cultivars are native to China (Table 1). ‘Luo Tian Tian Shi’ is included in the analysis.
The collected leaves were frozen and powdered with liquid nitrogen for total DNA extraction. Total DNA was extracted by the CTAB method of Doyle and Doyle (1987), followed by additional phenol extraction, and PEG (MW 8000) precipitation for DNA purification. The amount of DNA was calculated by measuring absorbance at 260 nm (Birren et al. 1997).
Amplified fragment length polymorphism analysis
AFLP analysis was performed using the AFLP Analysis System I Kit (Life Technologies, USA), following the manufacturer’s instructions. Briefly, 250 ng of total DNA was digested with 2.5 units of EcoRI and MseI. The DNA fragments were ligated to EcoRI and MseI adapters. After the pre-amplification reaction with the pre-amp primer mix, selective amplification was performed with six combinations of Label + EcoRI + 3 and MseI + 3 primers: FAM-E-AGC + M-CTA, HEX-E-ACC + M-CTA, NED-E-ACG + M-CAG, FAM-E-AGC + M-CAC, HEX-E-ACC + M-CAG, or NED-E-ACG + M-CTA. The amplified products were separated on an ABI PRISM 310 Genetic Analyzer (PE Biosystems, USA) after size standards were added.
Data analysis
AFLP bands were scored as present (1) or absent (0) and referenced to size standards. A total of 496 bands were scored. PIC values were calculated by the method of Roldan-Ruiz et al. (2000). After generation of a distance matrix (Nei and Li 1979), a multidimensional scaling analysis was conducted with NTSYS 2.0 (Rohlf 1998). TREECON (Van de Peer and Wachter 1994) was used to perform a neighbor-joining (NJ) analysis with 500 bootstrap replications conducted to establish statistical support levels. A consensus tree with support levels >50 % was generated (Fig. 1). Yonemori et al. (2008b) previously presented a preliminary NJ tree from some of this data, but without branch lengths and statistical support levels. Analysis of molecular variance (AMOVA) analysis and F st calculations were performed with Arlequin 3.1 (Excoffier et al. 2005). STRUCTURE 2.3 software was used to perform population structure analysis (Falush et al. 2003; Pritchard et al. 2000, 2007). The STRUCTURE model assumed admixture, hexaploid genetics, with missing bands coded as 0. The recessive allele model was implemented. Initially, test runs were conducted for 105 cycles after 104 burn-in cycles. F st, α, and ln Pr(X/K) were evaluated across the runs to determine whether longer runs were needed. All of the parameters were stable after the initial burn-in period. Duplicate runs of 5 × 104 to 105 cycles were run from K = 2 to K = 11. A set of five runs, each, was conducted from K = 2 to K = 7, with and without using prior population data as an additional evaluation tool. α values were considerably lower for the analyses without the prior population data. For the final analysis, duplicate runs were conducted with a burn-in of 2 × 104 followed by 5 × 105 cycles.
Results
Amplified fragment length polymorphism analysis
Four hundred ninety-six AFLP markers were obtained for each of the initial 153 cultivars. Four hundred sixty-three markers were polymorphic among the 146 cultivars retained for subsequent analysis. Mean marker PIC value was 0.185. Supplemental file 1 contains the AFLP matrix for all of the scored AFLPs.
Neighbor joining
Cultivars appeared to be primarily delineated by their country of origin in the NJ tree and secondarily by classification of astringency status. However, statistical support is <50 % for the short basal branches of the tree which separate cultivars by country of origin and astringency status. While most cultivars are clearly different at many loci, genetic differences that could be ascribed to specific geographic origins or astringency status could not be statistically proven by NJ. Several cultivars were placed in anomalous positions in the NJ Tree. ‘Luo Tian Tian Shi’ is placed with all of the other Chinese cultivars, but is a PCNA type, and might be expected to cluster with PCNA Japanese cultivars. However, the ‘Luo Tian Tian Shi’ PCNA mechanism (gene) is different than is found in Japanese cultivars (Ikegami et al. 2006). Two Korean cultivars, #121 ‘Goryo-soo-si,’ a PVA type, and #120 ‘Sa-gock-si’ did not cluster with the rest of the Korean cultivars. ‘Goryo-soo-si’ was associated with a group that had both PVNA and PCA types while ‘Sa-gock-si,’ a PCA type, was associated with PVNA Japanese cultivars. These cultivars may be misidentified as were the seven cultivars removed prior to statistical analysis.
Multidimensional scaling
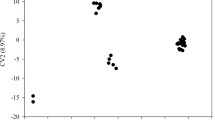
Multidimensional scaling (MDS) provided a different impression of variation among cultivars than observed in the NJ analysis. Two distinct clusters were obtained for the first three dimensions. Chinese and Korean cultivars clustered together but formed two subpopulations shown in Fig. 2 by country of origin; all of the Japanese cultivars formed a separate group. Within the Japanese population, there did not appear to be any significant differentiation between PCNA and non-PCNA cultivar types. The anomalous Korean cultivar, #121 Goryo-soo-si,’ was an outlier in the Korean group as were #122 and #123. However, #120 ‘Sa-gock-si’ clustered as expected with the other Korean cultivars.
Multidimensional scaling analysis for 146 persimmon cultivars. First three dimensions are shown. Country of origin is indicated by color, and numbers are referenced to the cultivar lists in Table 1. Three general groupings are indicated, Japanese cultivars and overlapping Korean-Chinese groups.
Analysis of molecular variance and F st estimation
Maximum likelihood AMOVA estimates were significant, P = 0.000, among cultivars by origin and in a separate analysis, among pollination types. However, in both analyses, most of the variation was found among cultivars, rather than among classification groups. Only 12.4 and 7.42 % of the total variation was partitioned into country or astringency classification, respectively, in the separate analyses (Table 2). Individual F sts for Japanese, Chinese, and Korean cultivars were similar, but low (Table 3). F sts for PCA, PCNA, PVA, and PVNA groups were also low but similar. Pairwise diversity comparisons for country and for astringency classification type were significant but low, since most polymorphism was partitioned among cultivars, not cultivar classes.
STRUCTURE analysis
Extensive testing of burn-in parameters and run cycles from K = 2 to K = 11 showed that α reached a minimum value of about α = 0.2 for K = 4 and higher (Fig. 3), slightly lower at K = 6. Mean F st values/population (within population heterogeneity values) was minimized from K = 4 to K = 7. Therefore, K values from 4 to 7 provided the most congruent arrangement of cultivars with country and astringency classification type (Fig. 4). However, statistics for K = 4 through K = 7 are similar, and there is no clear ‘best’ K. K = 6 is most congruent with our knowledge of cultivar country of origin. Korean and Chinese cultivars were quite distinct from the both the Japanese PCNA and Japanese non-PCNA types. Two subgroups appear to be present in the non-PCNA types, potentially representing the PVNA and PCA types which are the primary classes found in the non-PCNA population. These groups and a third PVA group are also seen in the NJ tree, but with poor basal branch support. There is some evidence of two distinct populations within the Japanese PCNA population at K = 5 through K = 7, something that we did not observe in the other clustering analyses. Even within the a priori cultivar groups, considerable within cultivar heterogeneity from other groups was observed for all K values, since most of the variation among these cultivars is between individuals rather than populations. Korean or Chinese cultivars comprised single classification groups in STRUCTURE.
Graphical output from STRUCTURE analysis of 496 markers from 146 persimmon cultivars. Clustering distributions for K values from K = 2 to 7 are shown (top to bottom). Cultivars are organized by country location. Group 1 = PCNA cultivars from Japan, group 2 = non-PCNA cultivars from Japan, group 3 = Korean cultivars, and group 4 = Chinese cultivars
Discussion
Naval et al. (2010) conducted the most extensive previous analysis of persimmon diversity using 71 cultivars from Europe and Japan, surveyed with 206 distinct polymorphisms from SSR markers. Their PCA, NJ, and AMOVA analyses were similar to the results from the present study, but their conclusions were different.
Examination of the AMOVAs from both studies showed that variation was found among cultivars of all types and was not due to cultivar origin or astringency classification type. We found that only 12.4 or 7.4 % of total variation could be explained by country of origin or astringency classification type, respectively, while Naval et al. (2010) found that 15 and 27 % of the variation in their study could be explained by country of origin and astringency classification type. The different results that they obtained for classification type are probably the result of differences in the populations being surveyed. A high proportion of random variation was present among cultivars in both studies.
The general form of the cladogram from Naval et al. (2010), computed with different software, was very similar to our NJ cladogram with strongly supported terminal clades with long branches, but poorly defined and supported basal groups separating the clades. Both studies produced cladograms that separated cultivars into distinct clades by astringency classification type or country of origin. The shape of the NJ tree separates PVA and PCNA among Japanese cultivars. Closer examination showed that the furcations separating cultivars by country or astringency type are not supported at the >50 % support level. The branches are short, reflecting the lack of diversity between groups.
Our MDS analysis, similar to the PCA analysis performed by Naval et al. (2010), suggested that there are differences due to cultivar origin. This is probably the result of relationship by descent as most of the Asian cultivars are relatively old and have less introgression than is found among European cultivars (Badenes et al. 2003; Yonemori et al. 2008a). Among the more ancient Asian cultivars, astringency classification type is highly coincident with cultivar origin (Table 1). European cultivars show a lower level of coincidence between astringency classification type and present cultivar location by nation (Yonemori et al. 2008a; Naval et al. 2010), due to recent selection of these cultivars from ancestral Asian groups. No significant clustering for astringency classification type (PCNA and PVA) was seen in the Japanese population from the MDS analysis.
Most of the molecular variation from AMOVA could not be ascribed to astringency classification type or country of origin. However, the results from the structure analysis showed that there are clusters within classes arranged by cultivar origin. These clusters cannot be fully explained by differences in pollination status and could represent subpopulation isolation or identity by descent.
Although molecular markers were not especially helpful for determining the history of modern cultivars due to strong admixture, some of these markers or marker combinations may be used for selection of genotypes with a desired astringency classification type. PCNA has been shown to be associated with a single locus, and absent recombination in the region adjacent to the locus, flanking markers should provide a means of selecting for PCNA at the seed/seedling stage (Kanzaki et al. 2010).
Conclusions
Chinese, Korean, and Japanese cultivars were separated by all analyses, and PCNA cultivars could be separated from non-PCNA cultivars, but support for these groups as defined by clustering algorithms and AMOVA was weak. All of the cultivar groups were similar at the molecular level with most polymorphism due to individual cultivar differences. The primary character separating them appears to be country of origin. Classification type by astringency is confounded with cultivar origin. When the effects of cultivar origin were removed, significant AFLP associations with classification type by astringency were not observed. Individual cultivars are very genetically distinct. While this may not be expected for wild populations of an outcrossing species, these cultivars are highly selected for quality traits and human intervention provides an opportunity for strong selection and cultivar differentiation.
References
Agricultural Research Station (1912) Investigation on persimmon cultivars (in Japanese). Bull Agric Res Stat (extra) 28:1–46
Akagi T, Kanzaki S, Gao M, Tao R, Parfitt DE, Yonemori K (2009) Quantitative real-time PCR to determine allele number for the astringency locus by analysis of a linked marker in Diospyros kaki Thunb. Tree Genet Genomes 5:483–492
Akagi T, Takeda Y, Yonemori K, Ikegami A, Kono A, Yamada M, Kanzaki S (2010) Quantitative genotyping for the astringency locus in hexaploid persimmon cultivars using quantitative real-time PCR. J Am Soc Hort Sci 135:59–66
Akagi T, Tao R, Tsujimoto T, Kono A, Yonemori K (2012) Fine genotyping of a highly polymorphic ASTRINGENCY-linked locus reveals variable hexasomic inheritance in persimmon (Diospyros kaki Thunb) cultivars. Tree Genet Genomes 8:195–204
Badenes M, Garces A, Romero C, Romero M, Clave J, Rovira M, Llacer G (2003) Genetic diversity of introduced and local Spanish persimmon cultivars revealed by RAPD markers. Genet Resour Crop Evol 50:579–585
Birren B, Green ED, Klapholz S, Myers RM, Roskams J (1997) Genome analysis: a laboratory manual 1:624
Cho SK, Cho TH (1965) Studies on the local varieties of persimmon in Korea Research Reports of the Office of Rural Development (Suweon). 8:147–190
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue Phytochemical Bulletin No 19. The Phytochemical Section of the Botanical Society of America, Irvine, pp 11–15
Du X, Zhang Q, Luo Z (2009) Identification of a Chinese PVNA type of Japanese persimmon discovered from Dabieshan region in central China. Acta Horticult 833:97–102
Excoffier L, Laval G, Schneider S (2005) Arlequin (version 3.0): an integrated software package for population genetics data analysis Evolutionary. Bioinformatics 1:47–50
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587
Guo DL, Luo ZR (2006) Genetic relationships of some PCNA persimmons (Diospyros kaki Thumb) from China and Japan revealed by SRAP analysis. Genet Resour Crop Evol 53:1597–1603
Guo DL, Luo ZR (2011) Genetic relationships of the Japanese persimmon Diospyros kaki (Ebenaceae) and related species revealed by SSR analysis. Genet Mol Res 10:1060–1068
Hume H (1913) Effect of pollination on the fruit of Diospyros kaki. Proc Am Soc Hort Sci 10:88–93
Hume HH (1914) A kaki classification. J Hered 5:400–406
Ikegami A, Eguchi S, Yonemori K, Yamada M, Sato A, Mitani N, Kitajima A (2006) Segregations of astringent progenies in the F1 populations derived from crosses between a Chinese pollination-constant nonastringent (PCNA) ‘Luo Tian Tian Shi’, and Japanese PCNA and pollination-constant astringent (PCA) cultivars of Japanese origin. Hortic Sci 41:561–563
Ikegami A, Eguchi S, Takashi A, Sato A, Yamada M, Kanzaki S, Kitajima A, Yonemori K (2011) Development of molecular markers linked to the allele associated with the non-astringent trait of the Chinese persimmon (Diospyros kaki Thunb). J Jpn Soc Hortic Sci 80:150–155
Kajiura (1946) Persimmon cultivars and their improvement 2 (in Japanese). Breed Hortic 1:175–182
Kanzaki S, Yonemori K, Sato A, Yamada M, Sugiura A (2000) Analysis of the genetic relationships among pollination-constant and non-astringent (PCNA) cultivars of persimmon (Diospyros kaki Thunb) from Japan and China using amplified fragment length polymorphism (AFLP). J Jpn Soc Hortic Sci 69:665–670
Kanzaki S, Yonemori K, Sugiura A, Sato A, Yamada M (2001) Identification of molecular markers linked to the trait of natural astringency loss of Japanese persimmon (Diospyros kaki) fruit. J Am Soc Hortic Sci 126:51–55
Kanzaki S, Akagi T, Masuko T, Kimura M, Yamada M, Sato A, Mitani N, Utsunomiya N, Yonemori K (2010) SCAR markers for practical application of marker-assisted selection in persimmon (Diospyros kaki Thunb) breeding. J Jpn Soc Hortic Sci 79:150–155
Kikuchi (1948) Pomology-part I (in Japanese). Yokendo, Tokyo, pp 347–400
Nei M, Li WH (1979) Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Soc USA 76:5269–5273
Naval MM, Zuriaga E, Pecchioli S, Llacer G, Giordani E, Badenes ML (2010) Analysis of genetic diversity among persimmon cultivars using microsatellite markers. Tree Genet Genomes 6:677–687
Pritchard JK, Stephens P, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Pritchard JK, Wen X, Falush D (2007) Documentation for structure software: version 2.2 Available from http://pritchardlab.stanford.edu/structure.html
Renzi W, Yong Y, Gaochao L (1997) Chinese persimmon germplasm resources. Acta Hortic 436:43–50
Rohlf FJ (1998) NTSYS-PC numerical taxonomy and multivariate analysis system, Version 2.0. Exeter Publications Setauket, New York
Roldan-Ruiz I, Dendauw J, Van Bockstaele E, Depicker A, DeLoose M (2000) AFLP markers reveal high polymorphic rates in ryegrasses (Lolium spp.). Mol Breed 6:125–134
Tamura M, Tao R, Yonemori K, Untunomia N, Sugiura A (1998) Ploidy level and genome size of several Diospyros species. J Jpn Soc Hortic Sci 67:306–312
van de Peer Y, De Wachter R (1994) TREECON for windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput Appl Biosci 10:569–570
Yamagishi M, Matsumoto S, Nakatuska A, Itamura H (2005) Identification of persimmon (Diospyros kaki) cultivars and phenetic relationships between Diospyros species by more effective RAPD analysis. Sci Hortic 105:283–290
Yonemori K, Honsho C, Kitajima A, Aradhya M, Giordani E, Bellini E, Parfitt DE (2008a) Relationship of European persimmon (Diospyros kaki Thunb) cultivars to Asian cultivars, characterized using AFLPs. Genet Resour Crop Evol 55:81–89
Yonemori K, Ikegami A, Kitajima A, Luo Z, Kanzaki S, Sato A, Yamada M, Yong Y, Wang R (2005) Existence of several pollination constant non-astringent type persimmon in China. Acta Hortic 685:77–83
Yonemori K, Kanzaki S, Honsho C, Akagi T, Parfitt DE (2008b) Phylogeny and cultivar development of Diospyros kaki: a survey based on molecular analyses. Adv Hortic Sci 22:261–268
Yonemori K, Sugiura A, Yamada M (2000) Persimmon genetics and breeding. In: J Janick (ed) Plant Breeding Rev. Wiley, New York 19:191-225
Acknowledgments
The technical assistance of Ms. Helen Chan is gratefully acknowledged. Research was supported in part by funds from the University of California Agricultural Experiment Station.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by D. Chagné
Electronic supplementary material
AFLP band scores for 146 persimmon cultivars used for statistical analyses are provided in supplemental file 1.
ESM 1
(XLSX 240 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Parfitt, D.E., Yonemori, K., Honsho, C. et al. Relationships among Asian persimmon cultivars, astringent and non-astringent types. Tree Genetics & Genomes 11, 24 (2015). https://doi.org/10.1007/s11295-015-0848-z
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11295-015-0848-z